
Microbulbifer sp. A4B17
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Microbulbiferaceae; Microbulbifer; unclassified Microbulbifer
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
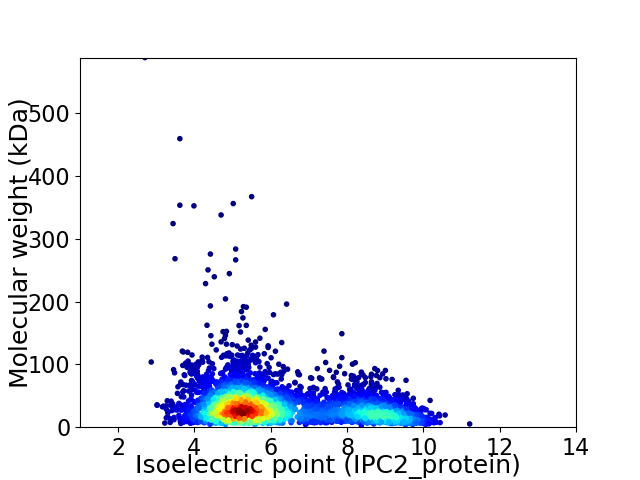
Virtual 2D-PAGE plot for 4240 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S1JNE0|A0A2S1JNE0_9GAMM Long-chain fatty acid--CoA ligase OS=Microbulbifer sp. A4B17 OX=359370 GN=BTJ40_03330 PE=4 SV=1
MM1 pKa = 7.27NFSLSSKK8 pKa = 10.0VRR10 pKa = 11.84FPLKK14 pKa = 10.3PIAAVVLFFCASNEE28 pKa = 3.89AFSAPGEE35 pKa = 4.07LSDD38 pKa = 4.02VPLYY42 pKa = 11.04SRR44 pKa = 11.84GSAEE48 pKa = 4.0PNLMFVLDD56 pKa = 4.3SSGSMNEE63 pKa = 3.69MVLPVDD69 pKa = 5.35GYY71 pKa = 10.18DD72 pKa = 3.34TSDD75 pKa = 3.6PAGSSCSSPISMTYY89 pKa = 9.68TSNSCRR95 pKa = 11.84GGGSCTRR102 pKa = 11.84YY103 pKa = 10.11YY104 pKa = 11.32DD105 pKa = 3.74DD106 pKa = 5.5VEE108 pKa = 4.59YY109 pKa = 10.85QVDD112 pKa = 3.73TSDD115 pKa = 3.38GSVDD119 pKa = 2.99IKK121 pKa = 11.21LVDD124 pKa = 3.62RR125 pKa = 11.84NRR127 pKa = 11.84AEE129 pKa = 4.62SEE131 pKa = 3.71WFTWNSSSNCFDD143 pKa = 3.39EE144 pKa = 5.46DD145 pKa = 3.56EE146 pKa = 5.18NYY148 pKa = 10.75SAWLYY153 pKa = 10.43AASSSGGGSYY163 pKa = 8.82YY164 pKa = 8.57TTGSSSGYY172 pKa = 9.27YY173 pKa = 9.26RR174 pKa = 11.84EE175 pKa = 4.25TVTGHH180 pKa = 5.99FLNWFFSKK188 pKa = 10.48EE189 pKa = 4.02NSSGGYY195 pKa = 9.19EE196 pKa = 3.79ADD198 pKa = 3.74EE199 pKa = 5.5FKK201 pKa = 11.27DD202 pKa = 4.11DD203 pKa = 4.63DD204 pKa = 4.83DD205 pKa = 3.84NAARR209 pKa = 11.84SKK211 pKa = 11.03VDD213 pKa = 2.98IRR215 pKa = 11.84RR216 pKa = 11.84TDD218 pKa = 2.91IMIGAAQEE226 pKa = 4.19LVNGLDD232 pKa = 3.7DD233 pKa = 3.88VRR235 pKa = 11.84IGLMQFNSTSGARR248 pKa = 11.84LLFGLEE254 pKa = 4.09SLTDD258 pKa = 3.7SNRR261 pKa = 11.84TDD263 pKa = 2.99ILDD266 pKa = 4.03IIEE269 pKa = 5.82DD270 pKa = 3.6IDD272 pKa = 4.32ANGYY276 pKa = 7.28TPLAEE281 pKa = 4.15SFAGVGRR288 pKa = 11.84YY289 pKa = 8.56FISGYY294 pKa = 10.08EE295 pKa = 4.27GNNLTYY301 pKa = 9.92IDD303 pKa = 3.73STGAEE308 pKa = 4.2TQDD311 pKa = 3.39PGEE314 pKa = 5.03DD315 pKa = 2.9IFVGATSGNSTTTILDD331 pKa = 3.5WNNISAPDD339 pKa = 3.59NTIDD343 pKa = 4.04GGAIQYY349 pKa = 8.42YY350 pKa = 7.19CQKK353 pKa = 10.63SFMVALTDD361 pKa = 4.8GEE363 pKa = 4.34PTEE366 pKa = 5.33DD367 pKa = 4.1NSISDD372 pKa = 4.45DD373 pKa = 3.63LTGYY377 pKa = 10.91DD378 pKa = 4.25YY379 pKa = 11.68GCSSNSGGCTSDD391 pKa = 4.01EE392 pKa = 3.91MDD394 pKa = 4.22DD395 pKa = 3.99VVKK398 pKa = 10.73ALYY401 pKa = 10.58DD402 pKa = 3.28IDD404 pKa = 5.29LRR406 pKa = 11.84PDD408 pKa = 3.48LKK410 pKa = 10.56KK411 pKa = 9.18TDD413 pKa = 3.5GEE415 pKa = 4.28EE416 pKa = 4.01VVNNITSYY424 pKa = 10.81IIGFAEE430 pKa = 5.5DD431 pKa = 4.06GLSDD435 pKa = 3.7TDD437 pKa = 3.62LMKK440 pKa = 10.94NAGEE444 pKa = 4.02LGGGGVYY451 pKa = 8.77TASNATEE458 pKa = 3.94LQLTFNTIINNVQSIVGSSSSVSFNTSSLEE488 pKa = 3.86ANSAIFSASFDD499 pKa = 3.49SSAWSGDD506 pKa = 3.56LEE508 pKa = 4.57VWSLDD513 pKa = 3.41DD514 pKa = 4.38EE515 pKa = 5.09GNISSTSSWQASSEE529 pKa = 4.21LEE531 pKa = 4.5SISADD536 pKa = 3.14NRR538 pKa = 11.84VMLSYY543 pKa = 10.72RR544 pKa = 11.84GGAGVAFTASGLGTSGTDD562 pKa = 3.49HH563 pKa = 7.66ADD565 pKa = 3.71DD566 pKa = 5.21LNLDD570 pKa = 3.56IQSNAGVDD578 pKa = 3.61DD579 pKa = 3.97GRR581 pKa = 11.84ASDD584 pKa = 4.33RR585 pKa = 11.84IDD587 pKa = 3.63YY588 pKa = 10.91LRR590 pKa = 11.84GDD592 pKa = 3.53QTDD595 pKa = 3.56EE596 pKa = 3.86GTEE599 pKa = 3.85AGDD602 pKa = 3.38FRR604 pKa = 11.84VRR606 pKa = 11.84EE607 pKa = 4.1SRR609 pKa = 11.84LGDD612 pKa = 3.33IVSSTPVYY620 pKa = 10.37VGEE623 pKa = 4.85PSSSWSEE630 pKa = 4.12AEE632 pKa = 3.85FSEE635 pKa = 4.54ASSYY639 pKa = 11.83DD640 pKa = 3.35SFVSSNSSRR649 pKa = 11.84TPVVYY654 pKa = 10.55VGANDD659 pKa = 3.99GFLHH663 pKa = 6.54GFNANVSGADD673 pKa = 3.26AGKK676 pKa = 10.37EE677 pKa = 3.8LIAYY681 pKa = 9.44SPAALLSTTADD692 pKa = 3.47EE693 pKa = 4.81GLHH696 pKa = 6.2ALSSQYY702 pKa = 8.69YY703 pKa = 4.47THH705 pKa = 7.48KK706 pKa = 10.47YY707 pKa = 9.26YY708 pKa = 11.18VDD710 pKa = 3.73GTPTASDD717 pKa = 3.81AYY719 pKa = 9.94IDD721 pKa = 4.45GSWEE725 pKa = 4.05TVLVGGLGGGGKK737 pKa = 10.23GYY739 pKa = 10.65YY740 pKa = 9.98ALNITNPSDD749 pKa = 3.45FTEE752 pKa = 4.62ANAADD757 pKa = 3.12IVMWEE762 pKa = 4.51FTDD765 pKa = 4.23SDD767 pKa = 4.33NNNLGYY773 pKa = 9.25TFSRR777 pKa = 11.84PQIGRR782 pKa = 11.84MSNGEE787 pKa = 3.42WAAIFGNGYY796 pKa = 10.21NSSTGDD802 pKa = 3.02AGLFIVYY809 pKa = 10.22LDD811 pKa = 4.74GEE813 pKa = 4.46DD814 pKa = 3.57SSGNSHH820 pKa = 7.03IYY822 pKa = 10.32ISTEE826 pKa = 3.78TADD829 pKa = 3.73TDD831 pKa = 4.26DD832 pKa = 4.65KK833 pKa = 11.94NGMSTPAIVDD843 pKa = 3.37SDD845 pKa = 4.23LDD847 pKa = 3.78GTIDD851 pKa = 4.46RR852 pKa = 11.84IYY854 pKa = 11.23AGDD857 pKa = 3.56LKK859 pKa = 11.24GNMWAFDD866 pKa = 3.64VSKK869 pKa = 9.81TGSWDD874 pKa = 3.49VASSTTSPSPLFSVSGEE891 pKa = 4.39AITGAPLVARR901 pKa = 11.84NTDD904 pKa = 3.29NSSGASPNLLVAFGTGQYY922 pKa = 10.8LVDD925 pKa = 4.72SDD927 pKa = 4.65TSDD930 pKa = 3.43TNPGGFYY937 pKa = 10.54VVSDD941 pKa = 3.5NDD943 pKa = 3.77VFNLDD948 pKa = 3.39TDD950 pKa = 3.86NLEE953 pKa = 4.04EE954 pKa = 4.03RR955 pKa = 11.84TLTSEE960 pKa = 5.0VLTLDD965 pKa = 4.64DD966 pKa = 4.74GSTTILRR973 pKa = 11.84TVSGSEE979 pKa = 4.02FTWSDD984 pKa = 2.77KK985 pKa = 11.25SGWYY989 pKa = 8.84MPLQEE994 pKa = 5.2GSTASQDD1001 pKa = 2.97GGEE1004 pKa = 4.15RR1005 pKa = 11.84VITRR1009 pKa = 11.84PEE1011 pKa = 3.77LQNYY1015 pKa = 8.12VLFFNTLIPTGQVCAAGGYY1034 pKa = 8.08GWLMSVDD1041 pKa = 3.67VWTGLAPEE1049 pKa = 5.08DD1050 pKa = 4.81SVFDD1054 pKa = 4.32ANNDD1058 pKa = 3.54GVIDD1062 pKa = 4.14EE1063 pKa = 5.38DD1064 pKa = 4.25DD1065 pKa = 3.5YY1066 pKa = 11.84GYY1068 pKa = 10.87VGEE1071 pKa = 4.68MLSEE1075 pKa = 4.27SSPNEE1080 pKa = 3.84SGFLGTKK1087 pKa = 9.83QYY1089 pKa = 10.94TGTSDD1094 pKa = 4.17GEE1096 pKa = 4.19VHH1098 pKa = 5.95EE1099 pKa = 5.03RR1100 pKa = 11.84EE1101 pKa = 4.19VDD1103 pKa = 3.94FGDD1106 pKa = 3.65GDD1108 pKa = 3.68RR1109 pKa = 11.84AGRR1112 pKa = 11.84LSWEE1116 pKa = 4.41EE1117 pKa = 3.44ITPNN1121 pKa = 3.5
MM1 pKa = 7.27NFSLSSKK8 pKa = 10.0VRR10 pKa = 11.84FPLKK14 pKa = 10.3PIAAVVLFFCASNEE28 pKa = 3.89AFSAPGEE35 pKa = 4.07LSDD38 pKa = 4.02VPLYY42 pKa = 11.04SRR44 pKa = 11.84GSAEE48 pKa = 4.0PNLMFVLDD56 pKa = 4.3SSGSMNEE63 pKa = 3.69MVLPVDD69 pKa = 5.35GYY71 pKa = 10.18DD72 pKa = 3.34TSDD75 pKa = 3.6PAGSSCSSPISMTYY89 pKa = 9.68TSNSCRR95 pKa = 11.84GGGSCTRR102 pKa = 11.84YY103 pKa = 10.11YY104 pKa = 11.32DD105 pKa = 3.74DD106 pKa = 5.5VEE108 pKa = 4.59YY109 pKa = 10.85QVDD112 pKa = 3.73TSDD115 pKa = 3.38GSVDD119 pKa = 2.99IKK121 pKa = 11.21LVDD124 pKa = 3.62RR125 pKa = 11.84NRR127 pKa = 11.84AEE129 pKa = 4.62SEE131 pKa = 3.71WFTWNSSSNCFDD143 pKa = 3.39EE144 pKa = 5.46DD145 pKa = 3.56EE146 pKa = 5.18NYY148 pKa = 10.75SAWLYY153 pKa = 10.43AASSSGGGSYY163 pKa = 8.82YY164 pKa = 8.57TTGSSSGYY172 pKa = 9.27YY173 pKa = 9.26RR174 pKa = 11.84EE175 pKa = 4.25TVTGHH180 pKa = 5.99FLNWFFSKK188 pKa = 10.48EE189 pKa = 4.02NSSGGYY195 pKa = 9.19EE196 pKa = 3.79ADD198 pKa = 3.74EE199 pKa = 5.5FKK201 pKa = 11.27DD202 pKa = 4.11DD203 pKa = 4.63DD204 pKa = 4.83DD205 pKa = 3.84NAARR209 pKa = 11.84SKK211 pKa = 11.03VDD213 pKa = 2.98IRR215 pKa = 11.84RR216 pKa = 11.84TDD218 pKa = 2.91IMIGAAQEE226 pKa = 4.19LVNGLDD232 pKa = 3.7DD233 pKa = 3.88VRR235 pKa = 11.84IGLMQFNSTSGARR248 pKa = 11.84LLFGLEE254 pKa = 4.09SLTDD258 pKa = 3.7SNRR261 pKa = 11.84TDD263 pKa = 2.99ILDD266 pKa = 4.03IIEE269 pKa = 5.82DD270 pKa = 3.6IDD272 pKa = 4.32ANGYY276 pKa = 7.28TPLAEE281 pKa = 4.15SFAGVGRR288 pKa = 11.84YY289 pKa = 8.56FISGYY294 pKa = 10.08EE295 pKa = 4.27GNNLTYY301 pKa = 9.92IDD303 pKa = 3.73STGAEE308 pKa = 4.2TQDD311 pKa = 3.39PGEE314 pKa = 5.03DD315 pKa = 2.9IFVGATSGNSTTTILDD331 pKa = 3.5WNNISAPDD339 pKa = 3.59NTIDD343 pKa = 4.04GGAIQYY349 pKa = 8.42YY350 pKa = 7.19CQKK353 pKa = 10.63SFMVALTDD361 pKa = 4.8GEE363 pKa = 4.34PTEE366 pKa = 5.33DD367 pKa = 4.1NSISDD372 pKa = 4.45DD373 pKa = 3.63LTGYY377 pKa = 10.91DD378 pKa = 4.25YY379 pKa = 11.68GCSSNSGGCTSDD391 pKa = 4.01EE392 pKa = 3.91MDD394 pKa = 4.22DD395 pKa = 3.99VVKK398 pKa = 10.73ALYY401 pKa = 10.58DD402 pKa = 3.28IDD404 pKa = 5.29LRR406 pKa = 11.84PDD408 pKa = 3.48LKK410 pKa = 10.56KK411 pKa = 9.18TDD413 pKa = 3.5GEE415 pKa = 4.28EE416 pKa = 4.01VVNNITSYY424 pKa = 10.81IIGFAEE430 pKa = 5.5DD431 pKa = 4.06GLSDD435 pKa = 3.7TDD437 pKa = 3.62LMKK440 pKa = 10.94NAGEE444 pKa = 4.02LGGGGVYY451 pKa = 8.77TASNATEE458 pKa = 3.94LQLTFNTIINNVQSIVGSSSSVSFNTSSLEE488 pKa = 3.86ANSAIFSASFDD499 pKa = 3.49SSAWSGDD506 pKa = 3.56LEE508 pKa = 4.57VWSLDD513 pKa = 3.41DD514 pKa = 4.38EE515 pKa = 5.09GNISSTSSWQASSEE529 pKa = 4.21LEE531 pKa = 4.5SISADD536 pKa = 3.14NRR538 pKa = 11.84VMLSYY543 pKa = 10.72RR544 pKa = 11.84GGAGVAFTASGLGTSGTDD562 pKa = 3.49HH563 pKa = 7.66ADD565 pKa = 3.71DD566 pKa = 5.21LNLDD570 pKa = 3.56IQSNAGVDD578 pKa = 3.61DD579 pKa = 3.97GRR581 pKa = 11.84ASDD584 pKa = 4.33RR585 pKa = 11.84IDD587 pKa = 3.63YY588 pKa = 10.91LRR590 pKa = 11.84GDD592 pKa = 3.53QTDD595 pKa = 3.56EE596 pKa = 3.86GTEE599 pKa = 3.85AGDD602 pKa = 3.38FRR604 pKa = 11.84VRR606 pKa = 11.84EE607 pKa = 4.1SRR609 pKa = 11.84LGDD612 pKa = 3.33IVSSTPVYY620 pKa = 10.37VGEE623 pKa = 4.85PSSSWSEE630 pKa = 4.12AEE632 pKa = 3.85FSEE635 pKa = 4.54ASSYY639 pKa = 11.83DD640 pKa = 3.35SFVSSNSSRR649 pKa = 11.84TPVVYY654 pKa = 10.55VGANDD659 pKa = 3.99GFLHH663 pKa = 6.54GFNANVSGADD673 pKa = 3.26AGKK676 pKa = 10.37EE677 pKa = 3.8LIAYY681 pKa = 9.44SPAALLSTTADD692 pKa = 3.47EE693 pKa = 4.81GLHH696 pKa = 6.2ALSSQYY702 pKa = 8.69YY703 pKa = 4.47THH705 pKa = 7.48KK706 pKa = 10.47YY707 pKa = 9.26YY708 pKa = 11.18VDD710 pKa = 3.73GTPTASDD717 pKa = 3.81AYY719 pKa = 9.94IDD721 pKa = 4.45GSWEE725 pKa = 4.05TVLVGGLGGGGKK737 pKa = 10.23GYY739 pKa = 10.65YY740 pKa = 9.98ALNITNPSDD749 pKa = 3.45FTEE752 pKa = 4.62ANAADD757 pKa = 3.12IVMWEE762 pKa = 4.51FTDD765 pKa = 4.23SDD767 pKa = 4.33NNNLGYY773 pKa = 9.25TFSRR777 pKa = 11.84PQIGRR782 pKa = 11.84MSNGEE787 pKa = 3.42WAAIFGNGYY796 pKa = 10.21NSSTGDD802 pKa = 3.02AGLFIVYY809 pKa = 10.22LDD811 pKa = 4.74GEE813 pKa = 4.46DD814 pKa = 3.57SSGNSHH820 pKa = 7.03IYY822 pKa = 10.32ISTEE826 pKa = 3.78TADD829 pKa = 3.73TDD831 pKa = 4.26DD832 pKa = 4.65KK833 pKa = 11.94NGMSTPAIVDD843 pKa = 3.37SDD845 pKa = 4.23LDD847 pKa = 3.78GTIDD851 pKa = 4.46RR852 pKa = 11.84IYY854 pKa = 11.23AGDD857 pKa = 3.56LKK859 pKa = 11.24GNMWAFDD866 pKa = 3.64VSKK869 pKa = 9.81TGSWDD874 pKa = 3.49VASSTTSPSPLFSVSGEE891 pKa = 4.39AITGAPLVARR901 pKa = 11.84NTDD904 pKa = 3.29NSSGASPNLLVAFGTGQYY922 pKa = 10.8LVDD925 pKa = 4.72SDD927 pKa = 4.65TSDD930 pKa = 3.43TNPGGFYY937 pKa = 10.54VVSDD941 pKa = 3.5NDD943 pKa = 3.77VFNLDD948 pKa = 3.39TDD950 pKa = 3.86NLEE953 pKa = 4.04EE954 pKa = 4.03RR955 pKa = 11.84TLTSEE960 pKa = 5.0VLTLDD965 pKa = 4.64DD966 pKa = 4.74GSTTILRR973 pKa = 11.84TVSGSEE979 pKa = 4.02FTWSDD984 pKa = 2.77KK985 pKa = 11.25SGWYY989 pKa = 8.84MPLQEE994 pKa = 5.2GSTASQDD1001 pKa = 2.97GGEE1004 pKa = 4.15RR1005 pKa = 11.84VITRR1009 pKa = 11.84PEE1011 pKa = 3.77LQNYY1015 pKa = 8.12VLFFNTLIPTGQVCAAGGYY1034 pKa = 8.08GWLMSVDD1041 pKa = 3.67VWTGLAPEE1049 pKa = 5.08DD1050 pKa = 4.81SVFDD1054 pKa = 4.32ANNDD1058 pKa = 3.54GVIDD1062 pKa = 4.14EE1063 pKa = 5.38DD1064 pKa = 4.25DD1065 pKa = 3.5YY1066 pKa = 11.84GYY1068 pKa = 10.87VGEE1071 pKa = 4.68MLSEE1075 pKa = 4.27SSPNEE1080 pKa = 3.84SGFLGTKK1087 pKa = 9.83QYY1089 pKa = 10.94TGTSDD1094 pKa = 4.17GEE1096 pKa = 4.19VHH1098 pKa = 5.95EE1099 pKa = 5.03RR1100 pKa = 11.84EE1101 pKa = 4.19VDD1103 pKa = 3.94FGDD1106 pKa = 3.65GDD1108 pKa = 3.68RR1109 pKa = 11.84AGRR1112 pKa = 11.84LSWEE1116 pKa = 4.41EE1117 pKa = 3.44ITPNN1121 pKa = 3.5
Molecular weight: 119.73 kDa
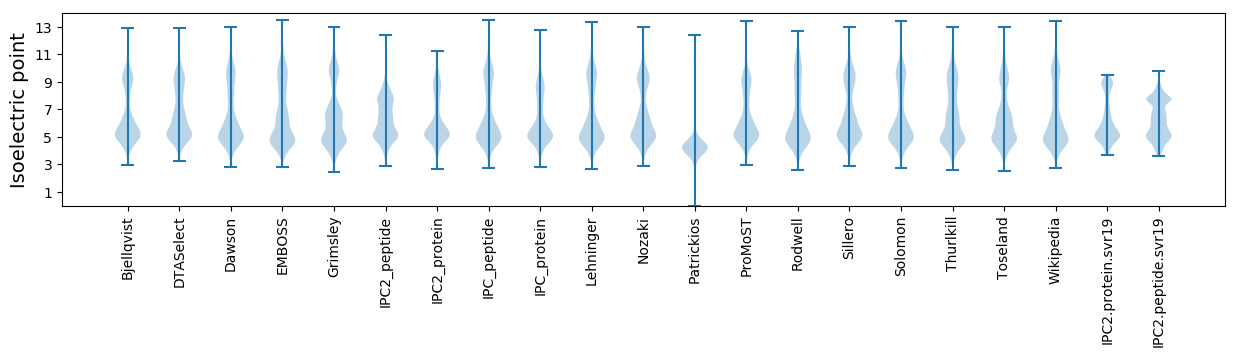
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S1JY68|A0A2S1JY68_9GAMM Sulfurtransferase TusA family protein OS=Microbulbifer sp. A4B17 OX=359370 GN=BTJ40_03770 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.2RR12 pKa = 11.84KK13 pKa = 7.57RR14 pKa = 11.84THH16 pKa = 5.9GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27NGRR28 pKa = 11.84KK29 pKa = 8.87ILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.2RR12 pKa = 11.84KK13 pKa = 7.57RR14 pKa = 11.84THH16 pKa = 5.9GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27NGRR28 pKa = 11.84KK29 pKa = 8.87ILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1368767 |
25 |
5896 |
322.8 |
35.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.873 ± 0.052 | 1.058 ± 0.016 |
5.661 ± 0.054 | 6.601 ± 0.041 |
4.008 ± 0.028 | 7.595 ± 0.043 |
2.07 ± 0.022 | 5.816 ± 0.031 |
4.415 ± 0.043 | 10.416 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.253 ± 0.022 | 3.94 ± 0.03 |
4.292 ± 0.031 | 4.176 ± 0.03 |
5.491 ± 0.045 | 6.966 ± 0.05 |
5.089 ± 0.049 | 6.742 ± 0.033 |
1.394 ± 0.019 | 3.146 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
