
Reinekea thalattae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Saccharospirillaceae; Reinekea
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
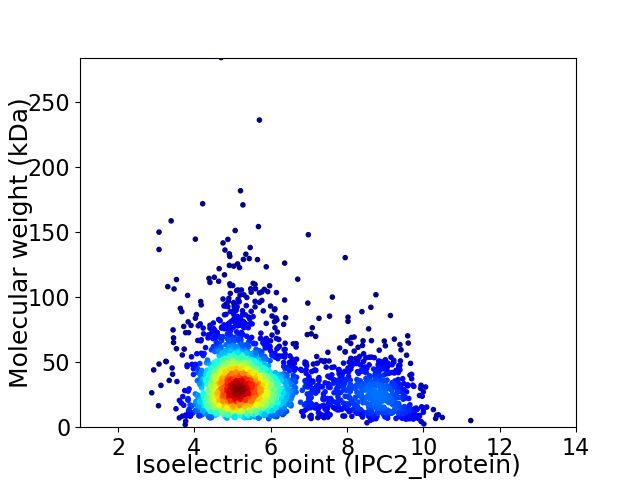
Virtual 2D-PAGE plot for 2651 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C8Z5F9|A0A5C8Z5F9_9GAMM VOC family protein OS=Reinekea thalattae OX=2593301 GN=FME95_01110 PE=4 SV=1
MM1 pKa = 8.42KK2 pKa = 10.04MMSVWVSALLALFVSACTSVNDD24 pKa = 4.07DD25 pKa = 3.95DD26 pKa = 4.82KK27 pKa = 11.93VVIGVATDD35 pKa = 3.35AEE37 pKa = 4.49KK38 pKa = 10.62QVLIHH43 pKa = 5.76YY44 pKa = 7.29LPWFADD50 pKa = 3.31EE51 pKa = 5.83NYY53 pKa = 9.28AQEE56 pKa = 4.37PSRR59 pKa = 11.84HH60 pKa = 5.04WSHH63 pKa = 5.58GTVNEE68 pKa = 4.13ALVGEE73 pKa = 4.56YY74 pKa = 10.33DD75 pKa = 3.75SQSWATQLYY84 pKa = 10.11HH85 pKa = 6.95ILLSAAVGVDD95 pKa = 2.78GAMINIRR102 pKa = 11.84TDD104 pKa = 3.19YY105 pKa = 11.38DD106 pKa = 3.5KK107 pKa = 11.47DD108 pKa = 3.44ALSIFMEE115 pKa = 4.08SLEE118 pKa = 4.21RR119 pKa = 11.84VRR121 pKa = 11.84AVYY124 pKa = 10.21PDD126 pKa = 3.87FAYY129 pKa = 10.45HH130 pKa = 6.53ISASIDD136 pKa = 3.56DD137 pKa = 3.86QDD139 pKa = 3.58KK140 pKa = 10.7TEE142 pKa = 4.89ALAQQEE148 pKa = 4.28FEE150 pKa = 4.81YY151 pKa = 10.55IAEE154 pKa = 4.94LIDD157 pKa = 4.17NYY159 pKa = 8.15TTYY162 pKa = 10.88LHH164 pKa = 7.04KK165 pKa = 10.76NGKK168 pKa = 8.44PVIFIWAYY176 pKa = 11.28NNLTPAQYY184 pKa = 10.64RR185 pKa = 11.84SIADD189 pKa = 3.27QVFGEE194 pKa = 4.64DD195 pKa = 4.29EE196 pKa = 4.57VILVQNDD203 pKa = 2.72IDD205 pKa = 3.92TSAVADD211 pKa = 4.99EE212 pKa = 4.5INMNAFYY219 pKa = 9.83PWVKK223 pKa = 10.51GFADD227 pKa = 5.07DD228 pKa = 4.21GSDD231 pKa = 3.03WGEE234 pKa = 4.17VYY236 pKa = 10.95LDD238 pKa = 3.07WFYY241 pKa = 10.99RR242 pKa = 11.84TSADD246 pKa = 3.41YY247 pKa = 9.26LTDD250 pKa = 3.41NKK252 pKa = 10.72QEE254 pKa = 4.37FIVAGVWPGFDD265 pKa = 3.23DD266 pKa = 5.47RR267 pKa = 11.84QVNWYY272 pKa = 9.76GDD274 pKa = 3.76DD275 pKa = 3.72YY276 pKa = 11.39AGRR279 pKa = 11.84WIDD282 pKa = 3.54RR283 pKa = 11.84ADD285 pKa = 3.22GAIYY289 pKa = 10.61DD290 pKa = 4.26NTWQLIDD297 pKa = 3.59TFNNEE302 pKa = 3.63WQQDD306 pKa = 3.25IATEE310 pKa = 4.8DD311 pKa = 3.39ISIDD315 pKa = 3.19WVVIEE320 pKa = 4.35TWNDD324 pKa = 3.13FNEE327 pKa = 4.58GSEE330 pKa = 4.22IEE332 pKa = 4.91PIAATADD339 pKa = 3.76TEE341 pKa = 4.44SFQYY345 pKa = 11.05LKK347 pKa = 10.26LTDD350 pKa = 3.73SYY352 pKa = 10.42ITNFKK357 pKa = 9.74ATDD360 pKa = 3.65SVLDD364 pKa = 4.43ADD366 pKa = 5.93DD367 pKa = 4.59EE368 pKa = 4.55LLNAAVKK375 pKa = 10.1IYY377 pKa = 10.25QAAKK381 pKa = 10.5LIDD384 pKa = 4.37DD385 pKa = 4.79GDD387 pKa = 3.94RR388 pKa = 11.84ASANYY393 pKa = 10.41YY394 pKa = 8.7STLEE398 pKa = 4.06QAIEE402 pKa = 4.31AYY404 pKa = 10.4LKK406 pKa = 10.8ADD408 pKa = 3.5AEE410 pKa = 4.52GASDD414 pKa = 3.88YY415 pKa = 11.59AEE417 pKa = 4.38TIIDD421 pKa = 3.74GG422 pKa = 3.98
MM1 pKa = 8.42KK2 pKa = 10.04MMSVWVSALLALFVSACTSVNDD24 pKa = 4.07DD25 pKa = 3.95DD26 pKa = 4.82KK27 pKa = 11.93VVIGVATDD35 pKa = 3.35AEE37 pKa = 4.49KK38 pKa = 10.62QVLIHH43 pKa = 5.76YY44 pKa = 7.29LPWFADD50 pKa = 3.31EE51 pKa = 5.83NYY53 pKa = 9.28AQEE56 pKa = 4.37PSRR59 pKa = 11.84HH60 pKa = 5.04WSHH63 pKa = 5.58GTVNEE68 pKa = 4.13ALVGEE73 pKa = 4.56YY74 pKa = 10.33DD75 pKa = 3.75SQSWATQLYY84 pKa = 10.11HH85 pKa = 6.95ILLSAAVGVDD95 pKa = 2.78GAMINIRR102 pKa = 11.84TDD104 pKa = 3.19YY105 pKa = 11.38DD106 pKa = 3.5KK107 pKa = 11.47DD108 pKa = 3.44ALSIFMEE115 pKa = 4.08SLEE118 pKa = 4.21RR119 pKa = 11.84VRR121 pKa = 11.84AVYY124 pKa = 10.21PDD126 pKa = 3.87FAYY129 pKa = 10.45HH130 pKa = 6.53ISASIDD136 pKa = 3.56DD137 pKa = 3.86QDD139 pKa = 3.58KK140 pKa = 10.7TEE142 pKa = 4.89ALAQQEE148 pKa = 4.28FEE150 pKa = 4.81YY151 pKa = 10.55IAEE154 pKa = 4.94LIDD157 pKa = 4.17NYY159 pKa = 8.15TTYY162 pKa = 10.88LHH164 pKa = 7.04KK165 pKa = 10.76NGKK168 pKa = 8.44PVIFIWAYY176 pKa = 11.28NNLTPAQYY184 pKa = 10.64RR185 pKa = 11.84SIADD189 pKa = 3.27QVFGEE194 pKa = 4.64DD195 pKa = 4.29EE196 pKa = 4.57VILVQNDD203 pKa = 2.72IDD205 pKa = 3.92TSAVADD211 pKa = 4.99EE212 pKa = 4.5INMNAFYY219 pKa = 9.83PWVKK223 pKa = 10.51GFADD227 pKa = 5.07DD228 pKa = 4.21GSDD231 pKa = 3.03WGEE234 pKa = 4.17VYY236 pKa = 10.95LDD238 pKa = 3.07WFYY241 pKa = 10.99RR242 pKa = 11.84TSADD246 pKa = 3.41YY247 pKa = 9.26LTDD250 pKa = 3.41NKK252 pKa = 10.72QEE254 pKa = 4.37FIVAGVWPGFDD265 pKa = 3.23DD266 pKa = 5.47RR267 pKa = 11.84QVNWYY272 pKa = 9.76GDD274 pKa = 3.76DD275 pKa = 3.72YY276 pKa = 11.39AGRR279 pKa = 11.84WIDD282 pKa = 3.54RR283 pKa = 11.84ADD285 pKa = 3.22GAIYY289 pKa = 10.61DD290 pKa = 4.26NTWQLIDD297 pKa = 3.59TFNNEE302 pKa = 3.63WQQDD306 pKa = 3.25IATEE310 pKa = 4.8DD311 pKa = 3.39ISIDD315 pKa = 3.19WVVIEE320 pKa = 4.35TWNDD324 pKa = 3.13FNEE327 pKa = 4.58GSEE330 pKa = 4.22IEE332 pKa = 4.91PIAATADD339 pKa = 3.76TEE341 pKa = 4.44SFQYY345 pKa = 11.05LKK347 pKa = 10.26LTDD350 pKa = 3.73SYY352 pKa = 10.42ITNFKK357 pKa = 9.74ATDD360 pKa = 3.65SVLDD364 pKa = 4.43ADD366 pKa = 5.93DD367 pKa = 4.59EE368 pKa = 4.55LLNAAVKK375 pKa = 10.1IYY377 pKa = 10.25QAAKK381 pKa = 10.5LIDD384 pKa = 4.37DD385 pKa = 4.79GDD387 pKa = 3.94RR388 pKa = 11.84ASANYY393 pKa = 10.41YY394 pKa = 8.7STLEE398 pKa = 4.06QAIEE402 pKa = 4.31AYY404 pKa = 10.4LKK406 pKa = 10.8ADD408 pKa = 3.5AEE410 pKa = 4.52GASDD414 pKa = 3.88YY415 pKa = 11.59AEE417 pKa = 4.38TIIDD421 pKa = 3.74GG422 pKa = 3.98
Molecular weight: 47.85 kDa
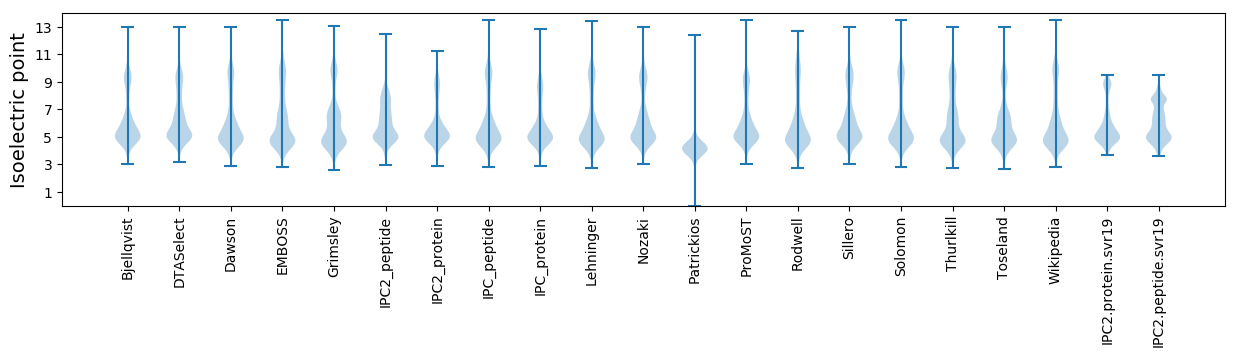
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C8ZBJ2|A0A5C8ZBJ2_9GAMM MATE family efflux transporter OS=Reinekea thalattae OX=2593301 GN=FME95_09855 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 8.92RR14 pKa = 11.84SHH16 pKa = 6.09GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.46NGRR28 pKa = 11.84KK29 pKa = 8.96VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84AALSAA44 pKa = 4.04
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 8.92RR14 pKa = 11.84SHH16 pKa = 6.09GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.46NGRR28 pKa = 11.84KK29 pKa = 8.96VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84AALSAA44 pKa = 4.04
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
861729 |
18 |
2531 |
325.1 |
36.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.235 ± 0.047 | 0.954 ± 0.015 |
5.662 ± 0.043 | 6.357 ± 0.045 |
4.043 ± 0.036 | 6.574 ± 0.04 |
2.113 ± 0.024 | 6.409 ± 0.039 |
4.792 ± 0.042 | 10.705 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.422 ± 0.025 | 4.076 ± 0.033 |
3.832 ± 0.031 | 5.042 ± 0.041 |
4.572 ± 0.036 | 6.932 ± 0.045 |
5.191 ± 0.039 | 6.832 ± 0.038 |
1.248 ± 0.018 | 3.009 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
