
Oceaniovalibus guishaninsula JLT2003
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Oceaniovalibus; Oceaniovalibus guishaninsula
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
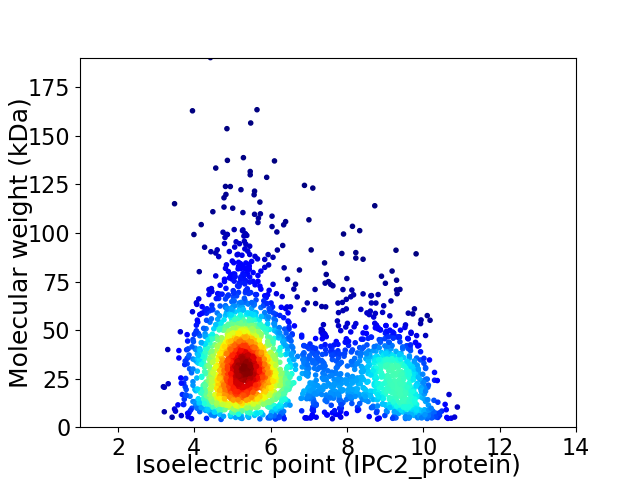
Virtual 2D-PAGE plot for 2840 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K2HF72|K2HF72_9RHOB PINc domain-containing protein OS=Oceaniovalibus guishaninsula JLT2003 OX=1231392 GN=OCGS_0817 PE=4 SV=1
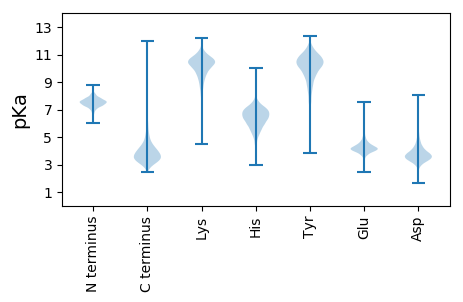
MM1 pKa = 7.23QRR3 pKa = 11.84TLYY6 pKa = 11.03ASVAALALGTGFAAAQDD23 pKa = 4.45LIFPVGEE30 pKa = 4.46GAFNWDD36 pKa = 3.7SYY38 pKa = 8.88QQFSDD43 pKa = 3.43ATDD46 pKa = 3.85LSGQTLDD53 pKa = 4.0ITGPWTGEE61 pKa = 3.96DD62 pKa = 3.23NDD64 pKa = 4.84LFTSVIAYY72 pKa = 9.97FEE74 pKa = 4.25EE75 pKa = 4.36ATGAEE80 pKa = 4.38VNYY83 pKa = 10.53SGSDD87 pKa = 3.18SFEE90 pKa = 3.74QDD92 pKa = 2.43IVISTRR98 pKa = 11.84SNSAPNIAVIPQPGLVGDD116 pKa = 4.8LASEE120 pKa = 5.5GYY122 pKa = 7.81LTPLAPEE129 pKa = 3.94TTDD132 pKa = 3.4WIVEE136 pKa = 4.13NYY138 pKa = 10.47AAGQSWADD146 pKa = 3.36LATFEE151 pKa = 4.68GQDD154 pKa = 3.6GQTQLYY160 pKa = 8.98AFPYY164 pKa = 9.67KK165 pKa = 9.85IDD167 pKa = 3.59VKK169 pKa = 10.85SLVWYY174 pKa = 10.29VPEE177 pKa = 3.88QFEE180 pKa = 4.17EE181 pKa = 3.99AGYY184 pKa = 10.35DD185 pKa = 3.5VPEE188 pKa = 4.18TLEE191 pKa = 4.0EE192 pKa = 3.99LKK194 pKa = 10.67QLNDD198 pKa = 3.44QIVADD203 pKa = 5.65GGTPWCIGLGSGAATGWPATDD224 pKa = 3.12WVEE227 pKa = 5.76DD228 pKa = 3.28MMLRR232 pKa = 11.84INPPEE237 pKa = 5.89IYY239 pKa = 10.11DD240 pKa = 3.3QWVSNDD246 pKa = 3.31IPFDD250 pKa = 4.19DD251 pKa = 4.67PQVVAAIEE259 pKa = 4.49EE260 pKa = 4.36YY261 pKa = 11.18GQFARR266 pKa = 11.84NDD268 pKa = 3.59NYY270 pKa = 10.77VSGGSSSVANTDD282 pKa = 4.16FRR284 pKa = 11.84DD285 pKa = 3.94APAGLFEE292 pKa = 5.6FPPQCYY298 pKa = 8.5MLKK301 pKa = 9.78QATFIPTFFPEE312 pKa = 4.29GTEE315 pKa = 3.84VGTDD319 pKa = 2.69VDD321 pKa = 4.82FFYY324 pKa = 11.12FPASAEE330 pKa = 4.04KK331 pKa = 10.83DD332 pKa = 3.03LGKK335 pKa = 9.73PVLGAGTMFTITKK348 pKa = 10.46DD349 pKa = 3.0SDD351 pKa = 3.23AAQAFIEE358 pKa = 4.34FLKK361 pKa = 10.39TPIAHH366 pKa = 6.39EE367 pKa = 3.85LWMAQSGFLTPYY379 pKa = 10.24KK380 pKa = 10.43DD381 pKa = 3.45ANTDD385 pKa = 3.47VFPSDD390 pKa = 3.35SQRR393 pKa = 11.84NMNDD397 pKa = 2.5ILTSATTFRR406 pKa = 11.84FDD408 pKa = 4.84GSDD411 pKa = 3.52LMPSEE416 pKa = 4.44IGAGAFWTGMVDD428 pKa = 3.45YY429 pKa = 7.62TTGADD434 pKa = 3.49AQTVASQIQSRR445 pKa = 11.84WEE447 pKa = 3.63QLKK450 pKa = 10.76
MM1 pKa = 7.23QRR3 pKa = 11.84TLYY6 pKa = 11.03ASVAALALGTGFAAAQDD23 pKa = 4.45LIFPVGEE30 pKa = 4.46GAFNWDD36 pKa = 3.7SYY38 pKa = 8.88QQFSDD43 pKa = 3.43ATDD46 pKa = 3.85LSGQTLDD53 pKa = 4.0ITGPWTGEE61 pKa = 3.96DD62 pKa = 3.23NDD64 pKa = 4.84LFTSVIAYY72 pKa = 9.97FEE74 pKa = 4.25EE75 pKa = 4.36ATGAEE80 pKa = 4.38VNYY83 pKa = 10.53SGSDD87 pKa = 3.18SFEE90 pKa = 3.74QDD92 pKa = 2.43IVISTRR98 pKa = 11.84SNSAPNIAVIPQPGLVGDD116 pKa = 4.8LASEE120 pKa = 5.5GYY122 pKa = 7.81LTPLAPEE129 pKa = 3.94TTDD132 pKa = 3.4WIVEE136 pKa = 4.13NYY138 pKa = 10.47AAGQSWADD146 pKa = 3.36LATFEE151 pKa = 4.68GQDD154 pKa = 3.6GQTQLYY160 pKa = 8.98AFPYY164 pKa = 9.67KK165 pKa = 9.85IDD167 pKa = 3.59VKK169 pKa = 10.85SLVWYY174 pKa = 10.29VPEE177 pKa = 3.88QFEE180 pKa = 4.17EE181 pKa = 3.99AGYY184 pKa = 10.35DD185 pKa = 3.5VPEE188 pKa = 4.18TLEE191 pKa = 4.0EE192 pKa = 3.99LKK194 pKa = 10.67QLNDD198 pKa = 3.44QIVADD203 pKa = 5.65GGTPWCIGLGSGAATGWPATDD224 pKa = 3.12WVEE227 pKa = 5.76DD228 pKa = 3.28MMLRR232 pKa = 11.84INPPEE237 pKa = 5.89IYY239 pKa = 10.11DD240 pKa = 3.3QWVSNDD246 pKa = 3.31IPFDD250 pKa = 4.19DD251 pKa = 4.67PQVVAAIEE259 pKa = 4.49EE260 pKa = 4.36YY261 pKa = 11.18GQFARR266 pKa = 11.84NDD268 pKa = 3.59NYY270 pKa = 10.77VSGGSSSVANTDD282 pKa = 4.16FRR284 pKa = 11.84DD285 pKa = 3.94APAGLFEE292 pKa = 5.6FPPQCYY298 pKa = 8.5MLKK301 pKa = 9.78QATFIPTFFPEE312 pKa = 4.29GTEE315 pKa = 3.84VGTDD319 pKa = 2.69VDD321 pKa = 4.82FFYY324 pKa = 11.12FPASAEE330 pKa = 4.04KK331 pKa = 10.83DD332 pKa = 3.03LGKK335 pKa = 9.73PVLGAGTMFTITKK348 pKa = 10.46DD349 pKa = 3.0SDD351 pKa = 3.23AAQAFIEE358 pKa = 4.34FLKK361 pKa = 10.39TPIAHH366 pKa = 6.39EE367 pKa = 3.85LWMAQSGFLTPYY379 pKa = 10.24KK380 pKa = 10.43DD381 pKa = 3.45ANTDD385 pKa = 3.47VFPSDD390 pKa = 3.35SQRR393 pKa = 11.84NMNDD397 pKa = 2.5ILTSATTFRR406 pKa = 11.84FDD408 pKa = 4.84GSDD411 pKa = 3.52LMPSEE416 pKa = 4.44IGAGAFWTGMVDD428 pKa = 3.45YY429 pKa = 7.62TTGADD434 pKa = 3.49AQTVASQIQSRR445 pKa = 11.84WEE447 pKa = 3.63QLKK450 pKa = 10.76
Molecular weight: 49.14 kDa
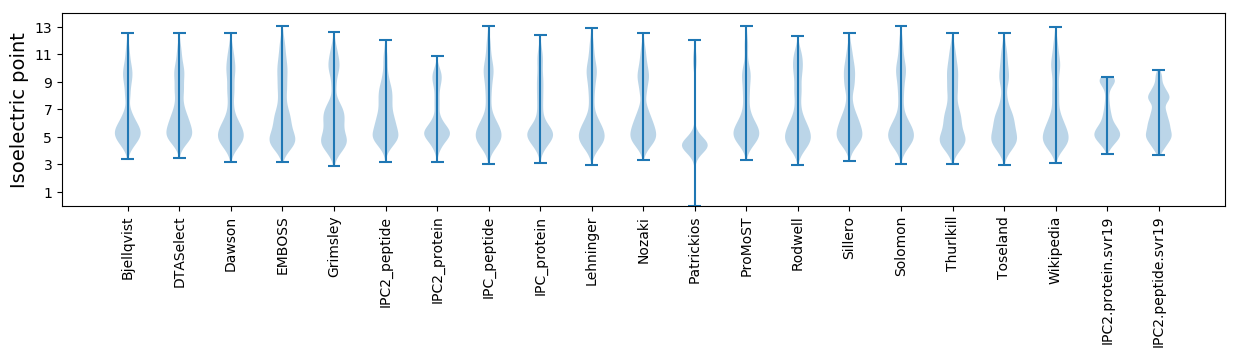
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K2HN72|K2HN72_9RHOB Pyrophosphate phospho-hydrolase OS=Oceaniovalibus guishaninsula JLT2003 OX=1231392 GN=OCGS_1820 PE=3 SV=1
MM1 pKa = 7.51TKK3 pKa = 9.12RR4 pKa = 11.84TFQPSNLVRR13 pKa = 11.84KK14 pKa = 8.83HH15 pKa = 4.64RR16 pKa = 11.84HH17 pKa = 3.91GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.37AGRR29 pKa = 11.84KK30 pKa = 8.54ILNARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.03GRR40 pKa = 11.84KK41 pKa = 8.43SLCAA45 pKa = 3.75
MM1 pKa = 7.51TKK3 pKa = 9.12RR4 pKa = 11.84TFQPSNLVRR13 pKa = 11.84KK14 pKa = 8.83HH15 pKa = 4.64RR16 pKa = 11.84HH17 pKa = 3.91GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.37AGRR29 pKa = 11.84KK30 pKa = 8.54ILNARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.03GRR40 pKa = 11.84KK41 pKa = 8.43SLCAA45 pKa = 3.75
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
883308 |
37 |
1797 |
311.0 |
33.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.666 ± 0.075 | 0.852 ± 0.015 |
6.808 ± 0.041 | 5.022 ± 0.046 |
3.491 ± 0.031 | 9.172 ± 0.043 |
1.974 ± 0.025 | 4.808 ± 0.033 |
2.22 ± 0.034 | 10.145 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.706 ± 0.023 | 2.152 ± 0.021 |
5.519 ± 0.032 | 2.848 ± 0.026 |
8.009 ± 0.049 | 4.397 ± 0.031 |
5.337 ± 0.03 | 7.397 ± 0.04 |
1.453 ± 0.022 | 2.023 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
