
Thiohalobacter thiocyanaticus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Chromatiales incertae sedis; Thiohalobacter
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
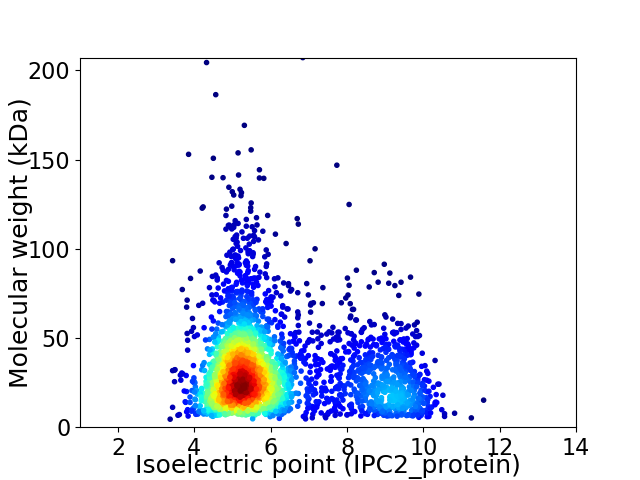
Virtual 2D-PAGE plot for 3024 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z4VRQ8|A0A1Z4VRQ8_9GAMM Cyclic nucleotide-binding protein OS=Thiohalobacter thiocyanaticus OX=585455 GN=FOKN1_1936 PE=4 SV=1
MM1 pKa = 7.81GIDD4 pKa = 4.11LPAAAAEE11 pKa = 4.28RR12 pKa = 11.84LRR14 pKa = 11.84HH15 pKa = 5.84HH16 pKa = 6.91PLVADD21 pKa = 3.94VEE23 pKa = 4.48QARR26 pKa = 11.84SFEE29 pKa = 4.14LHH31 pKa = 6.11AQQLPTGIDD40 pKa = 3.34RR41 pKa = 11.84INAEE45 pKa = 4.29LHH47 pKa = 6.19PGAGIDD53 pKa = 3.81GSEE56 pKa = 4.23SLVDD60 pKa = 3.41VDD62 pKa = 4.83IAIIDD67 pKa = 3.67TGVEE71 pKa = 4.14LNHH74 pKa = 7.34PDD76 pKa = 3.54LNVYY80 pKa = 9.68QYY82 pKa = 11.26AYY84 pKa = 10.01CYY86 pKa = 10.34QKK88 pKa = 9.4TPRR91 pKa = 11.84SGTCNTGDD99 pKa = 3.27SRR101 pKa = 11.84ANDD104 pKa = 3.56VQGHH108 pKa = 4.8GTHH111 pKa = 5.91VAGIAAARR119 pKa = 11.84DD120 pKa = 3.48NGIGVVGVAPGARR133 pKa = 11.84IWALNVFGTDD143 pKa = 3.24PGAGTGLLEE152 pKa = 4.96IIAALDD158 pKa = 3.63YY159 pKa = 11.24VIANAAQIEE168 pKa = 4.64VVNMSLGFTHH178 pKa = 7.74DD179 pKa = 3.57GTGSRR184 pKa = 11.84NFNEE188 pKa = 5.64AIDD191 pKa = 3.83NTLAAGITVVTSAGNDD207 pKa = 3.99AIDD210 pKa = 4.03VMYY213 pKa = 9.68TSPADD218 pKa = 3.45HH219 pKa = 7.26PGVITVSALADD230 pKa = 3.49GDD232 pKa = 3.75GRR234 pKa = 11.84IGARR238 pKa = 11.84GSLGFTYY245 pKa = 9.21ATADD249 pKa = 3.63GKK251 pKa = 9.61CTEE254 pKa = 4.32KK255 pKa = 10.75QDD257 pKa = 5.16DD258 pKa = 4.03SFACFSNYY266 pKa = 7.91GTGVDD271 pKa = 3.38IMAPGVEE278 pKa = 4.01IHH280 pKa = 5.98STYY283 pKa = 11.03LDD285 pKa = 3.69DD286 pKa = 5.93DD287 pKa = 4.14YY288 pKa = 11.49EE289 pKa = 4.39YY290 pKa = 11.82LHH292 pKa = 6.2GTSMASPHH300 pKa = 5.49VAGAAALYY308 pKa = 9.97LAANPGAGPADD319 pKa = 3.54VKK321 pKa = 11.01AALIAMGDD329 pKa = 3.88PAPCGASLSLCGDD342 pKa = 3.94DD343 pKa = 4.28PDD345 pKa = 6.03GIQEE349 pKa = 4.3PLVLLAPHH357 pKa = 7.78PDD359 pKa = 3.36TDD361 pKa = 4.34ADD363 pKa = 4.13TVLDD367 pKa = 5.11DD368 pKa = 6.09LDD370 pKa = 4.1NCPDD374 pKa = 3.34VHH376 pKa = 8.04NPDD379 pKa = 3.19QADD382 pKa = 3.64RR383 pKa = 11.84DD384 pKa = 3.73GDD386 pKa = 4.34GIGDD390 pKa = 3.75VCDD393 pKa = 3.99SDD395 pKa = 5.6GDD397 pKa = 4.54GIDD400 pKa = 4.09NGSDD404 pKa = 3.36NCPDD408 pKa = 3.51TANADD413 pKa = 3.5QRR415 pKa = 11.84DD416 pKa = 3.54QDD418 pKa = 3.47GDD420 pKa = 4.36GIGDD424 pKa = 3.71ACDD427 pKa = 3.27NCILHH432 pKa = 6.99ANAPQRR438 pKa = 11.84DD439 pKa = 3.87TDD441 pKa = 3.47GDD443 pKa = 4.36GYY445 pKa = 11.27GNRR448 pKa = 11.84CDD450 pKa = 5.29ADD452 pKa = 3.93FNNDD456 pKa = 2.9GAVNFSDD463 pKa = 4.48LAYY466 pKa = 10.36LKK468 pKa = 10.77SAFFSTDD475 pKa = 2.44PHH477 pKa = 7.47ADD479 pKa = 3.47LNGDD483 pKa = 3.65GAVNFADD490 pKa = 4.3LAILKK495 pKa = 10.03SFFFSTPGPSALDD508 pKa = 3.54PP509 pKa = 4.54
MM1 pKa = 7.81GIDD4 pKa = 4.11LPAAAAEE11 pKa = 4.28RR12 pKa = 11.84LRR14 pKa = 11.84HH15 pKa = 5.84HH16 pKa = 6.91PLVADD21 pKa = 3.94VEE23 pKa = 4.48QARR26 pKa = 11.84SFEE29 pKa = 4.14LHH31 pKa = 6.11AQQLPTGIDD40 pKa = 3.34RR41 pKa = 11.84INAEE45 pKa = 4.29LHH47 pKa = 6.19PGAGIDD53 pKa = 3.81GSEE56 pKa = 4.23SLVDD60 pKa = 3.41VDD62 pKa = 4.83IAIIDD67 pKa = 3.67TGVEE71 pKa = 4.14LNHH74 pKa = 7.34PDD76 pKa = 3.54LNVYY80 pKa = 9.68QYY82 pKa = 11.26AYY84 pKa = 10.01CYY86 pKa = 10.34QKK88 pKa = 9.4TPRR91 pKa = 11.84SGTCNTGDD99 pKa = 3.27SRR101 pKa = 11.84ANDD104 pKa = 3.56VQGHH108 pKa = 4.8GTHH111 pKa = 5.91VAGIAAARR119 pKa = 11.84DD120 pKa = 3.48NGIGVVGVAPGARR133 pKa = 11.84IWALNVFGTDD143 pKa = 3.24PGAGTGLLEE152 pKa = 4.96IIAALDD158 pKa = 3.63YY159 pKa = 11.24VIANAAQIEE168 pKa = 4.64VVNMSLGFTHH178 pKa = 7.74DD179 pKa = 3.57GTGSRR184 pKa = 11.84NFNEE188 pKa = 5.64AIDD191 pKa = 3.83NTLAAGITVVTSAGNDD207 pKa = 3.99AIDD210 pKa = 4.03VMYY213 pKa = 9.68TSPADD218 pKa = 3.45HH219 pKa = 7.26PGVITVSALADD230 pKa = 3.49GDD232 pKa = 3.75GRR234 pKa = 11.84IGARR238 pKa = 11.84GSLGFTYY245 pKa = 9.21ATADD249 pKa = 3.63GKK251 pKa = 9.61CTEE254 pKa = 4.32KK255 pKa = 10.75QDD257 pKa = 5.16DD258 pKa = 4.03SFACFSNYY266 pKa = 7.91GTGVDD271 pKa = 3.38IMAPGVEE278 pKa = 4.01IHH280 pKa = 5.98STYY283 pKa = 11.03LDD285 pKa = 3.69DD286 pKa = 5.93DD287 pKa = 4.14YY288 pKa = 11.49EE289 pKa = 4.39YY290 pKa = 11.82LHH292 pKa = 6.2GTSMASPHH300 pKa = 5.49VAGAAALYY308 pKa = 9.97LAANPGAGPADD319 pKa = 3.54VKK321 pKa = 11.01AALIAMGDD329 pKa = 3.88PAPCGASLSLCGDD342 pKa = 3.94DD343 pKa = 4.28PDD345 pKa = 6.03GIQEE349 pKa = 4.3PLVLLAPHH357 pKa = 7.78PDD359 pKa = 3.36TDD361 pKa = 4.34ADD363 pKa = 4.13TVLDD367 pKa = 5.11DD368 pKa = 6.09LDD370 pKa = 4.1NCPDD374 pKa = 3.34VHH376 pKa = 8.04NPDD379 pKa = 3.19QADD382 pKa = 3.64RR383 pKa = 11.84DD384 pKa = 3.73GDD386 pKa = 4.34GIGDD390 pKa = 3.75VCDD393 pKa = 3.99SDD395 pKa = 5.6GDD397 pKa = 4.54GIDD400 pKa = 4.09NGSDD404 pKa = 3.36NCPDD408 pKa = 3.51TANADD413 pKa = 3.5QRR415 pKa = 11.84DD416 pKa = 3.54QDD418 pKa = 3.47GDD420 pKa = 4.36GIGDD424 pKa = 3.71ACDD427 pKa = 3.27NCILHH432 pKa = 6.99ANAPQRR438 pKa = 11.84DD439 pKa = 3.87TDD441 pKa = 3.47GDD443 pKa = 4.36GYY445 pKa = 11.27GNRR448 pKa = 11.84CDD450 pKa = 5.29ADD452 pKa = 3.93FNNDD456 pKa = 2.9GAVNFSDD463 pKa = 4.48LAYY466 pKa = 10.36LKK468 pKa = 10.77SAFFSTDD475 pKa = 2.44PHH477 pKa = 7.47ADD479 pKa = 3.47LNGDD483 pKa = 3.65GAVNFADD490 pKa = 4.3LAILKK495 pKa = 10.03SFFFSTPGPSALDD508 pKa = 3.54PP509 pKa = 4.54
Molecular weight: 52.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z4VM51|A0A1Z4VM51_9GAMM Uncharacterized protein OS=Thiohalobacter thiocyanaticus OX=585455 GN=FOKN1_0292 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.2RR12 pKa = 11.84KK13 pKa = 7.57RR14 pKa = 11.84THH16 pKa = 5.88GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 9.83NGRR28 pKa = 11.84RR29 pKa = 11.84VLKK32 pKa = 10.45ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.17GRR39 pKa = 11.84ARR41 pKa = 11.84LSVV44 pKa = 3.12
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.2RR12 pKa = 11.84KK13 pKa = 7.57RR14 pKa = 11.84THH16 pKa = 5.88GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 9.83NGRR28 pKa = 11.84RR29 pKa = 11.84VLKK32 pKa = 10.45ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.17GRR39 pKa = 11.84ARR41 pKa = 11.84LSVV44 pKa = 3.12
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
944797 |
43 |
1891 |
312.4 |
34.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.977 ± 0.06 | 0.936 ± 0.016 |
5.919 ± 0.034 | 6.654 ± 0.048 |
3.382 ± 0.03 | 8.133 ± 0.044 |
2.434 ± 0.021 | 4.888 ± 0.036 |
2.715 ± 0.04 | 11.308 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.325 ± 0.022 | 2.68 ± 0.025 |
5.061 ± 0.038 | 4.031 ± 0.034 |
7.832 ± 0.048 | 4.928 ± 0.036 |
4.796 ± 0.033 | 6.971 ± 0.041 |
1.377 ± 0.019 | 2.652 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
