
Rousettus aegyptiacus polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Betapolyomavirus
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
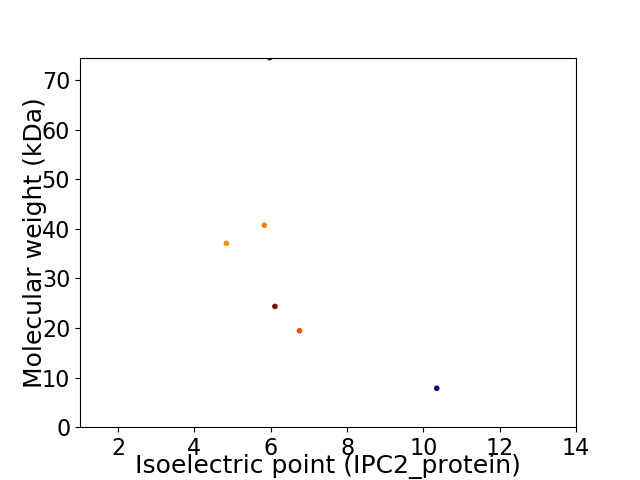
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S7J032|A0A1S7J032_9POLY Minor capsid protein OS=Rousettus aegyptiacus polyomavirus 1 OX=1904411 PE=3 SV=1
MM1 pKa = 7.31GALLAVLAEE10 pKa = 4.15VFEE13 pKa = 4.66ISAATGFTVDD23 pKa = 4.99SILTGEE29 pKa = 4.41ALVADD34 pKa = 5.08EE35 pKa = 5.43LLQAYY40 pKa = 7.15VTNLVTLEE48 pKa = 4.11GFTEE52 pKa = 4.83GEE54 pKa = 3.88ALLAIGFSPEE64 pKa = 3.46AAQLLTSLAPTFPEE78 pKa = 4.41AFTLVAGTEE87 pKa = 4.17ALVHH91 pKa = 6.19GSLFIGAATGAALAPYY107 pKa = 8.99SWDD110 pKa = 3.37YY111 pKa = 9.65ATPIADD117 pKa = 4.24LNQHH121 pKa = 6.35LMALQVWQPDD131 pKa = 4.11LNWEE135 pKa = 4.05DD136 pKa = 4.19VYY138 pKa = 11.37FPGVLPFARR147 pKa = 11.84FVNYY151 pKa = 9.56IDD153 pKa = 3.88PANWASNLYY162 pKa = 9.65HH163 pKa = 7.6AIGRR167 pKa = 11.84YY168 pKa = 7.45FWEE171 pKa = 4.21TAQRR175 pKa = 11.84AGTRR179 pKa = 11.84LIEE182 pKa = 3.87QEE184 pKa = 4.14VRR186 pKa = 11.84HH187 pKa = 5.78VSTDD191 pKa = 3.12LAQRR195 pKa = 11.84TVTSIAEE202 pKa = 3.8TLARR206 pKa = 11.84YY207 pKa = 9.22FEE209 pKa = 4.06NARR212 pKa = 11.84WAVSEE217 pKa = 4.48VTSGTYY223 pKa = 10.04GALQQYY229 pKa = 9.88YY230 pKa = 10.36SEE232 pKa = 5.11LPPLRR237 pKa = 11.84PPQIRR242 pKa = 11.84ALHH245 pKa = 5.79KK246 pKa = 10.64RR247 pKa = 11.84LGEE250 pKa = 4.3DD251 pKa = 3.06APSRR255 pKa = 11.84YY256 pKa = 10.41SFDD259 pKa = 4.06SNGNGSAQYY268 pKa = 10.32VDD270 pKa = 5.1KK271 pKa = 11.41VDD273 pKa = 3.61APGGARR279 pKa = 11.84QRR281 pKa = 11.84HH282 pKa = 4.9TPDD285 pKa = 2.34WMLPLILGLYY295 pKa = 9.58GDD297 pKa = 5.31IYY299 pKa = 10.35PSWEE303 pKa = 3.8ATLEE307 pKa = 3.98EE308 pKa = 4.93LEE310 pKa = 4.76AEE312 pKa = 3.99EE313 pKa = 5.87DD314 pKa = 4.02GPKK317 pKa = 9.98KK318 pKa = 10.3KK319 pKa = 10.28KK320 pKa = 9.6PRR322 pKa = 11.84SEE324 pKa = 3.67GRR326 pKa = 11.84RR327 pKa = 11.84TRR329 pKa = 11.84GRR331 pKa = 11.84KK332 pKa = 8.63SQSSSS337 pKa = 2.78
MM1 pKa = 7.31GALLAVLAEE10 pKa = 4.15VFEE13 pKa = 4.66ISAATGFTVDD23 pKa = 4.99SILTGEE29 pKa = 4.41ALVADD34 pKa = 5.08EE35 pKa = 5.43LLQAYY40 pKa = 7.15VTNLVTLEE48 pKa = 4.11GFTEE52 pKa = 4.83GEE54 pKa = 3.88ALLAIGFSPEE64 pKa = 3.46AAQLLTSLAPTFPEE78 pKa = 4.41AFTLVAGTEE87 pKa = 4.17ALVHH91 pKa = 6.19GSLFIGAATGAALAPYY107 pKa = 8.99SWDD110 pKa = 3.37YY111 pKa = 9.65ATPIADD117 pKa = 4.24LNQHH121 pKa = 6.35LMALQVWQPDD131 pKa = 4.11LNWEE135 pKa = 4.05DD136 pKa = 4.19VYY138 pKa = 11.37FPGVLPFARR147 pKa = 11.84FVNYY151 pKa = 9.56IDD153 pKa = 3.88PANWASNLYY162 pKa = 9.65HH163 pKa = 7.6AIGRR167 pKa = 11.84YY168 pKa = 7.45FWEE171 pKa = 4.21TAQRR175 pKa = 11.84AGTRR179 pKa = 11.84LIEE182 pKa = 3.87QEE184 pKa = 4.14VRR186 pKa = 11.84HH187 pKa = 5.78VSTDD191 pKa = 3.12LAQRR195 pKa = 11.84TVTSIAEE202 pKa = 3.8TLARR206 pKa = 11.84YY207 pKa = 9.22FEE209 pKa = 4.06NARR212 pKa = 11.84WAVSEE217 pKa = 4.48VTSGTYY223 pKa = 10.04GALQQYY229 pKa = 9.88YY230 pKa = 10.36SEE232 pKa = 5.11LPPLRR237 pKa = 11.84PPQIRR242 pKa = 11.84ALHH245 pKa = 5.79KK246 pKa = 10.64RR247 pKa = 11.84LGEE250 pKa = 4.3DD251 pKa = 3.06APSRR255 pKa = 11.84YY256 pKa = 10.41SFDD259 pKa = 4.06SNGNGSAQYY268 pKa = 10.32VDD270 pKa = 5.1KK271 pKa = 11.41VDD273 pKa = 3.61APGGARR279 pKa = 11.84QRR281 pKa = 11.84HH282 pKa = 4.9TPDD285 pKa = 2.34WMLPLILGLYY295 pKa = 9.58GDD297 pKa = 5.31IYY299 pKa = 10.35PSWEE303 pKa = 3.8ATLEE307 pKa = 3.98EE308 pKa = 4.93LEE310 pKa = 4.76AEE312 pKa = 3.99EE313 pKa = 5.87DD314 pKa = 4.02GPKK317 pKa = 9.98KK318 pKa = 10.3KK319 pKa = 10.28KK320 pKa = 9.6PRR322 pKa = 11.84SEE324 pKa = 3.67GRR326 pKa = 11.84RR327 pKa = 11.84TRR329 pKa = 11.84GRR331 pKa = 11.84KK332 pKa = 8.63SQSSSS337 pKa = 2.78
Molecular weight: 37.06 kDa
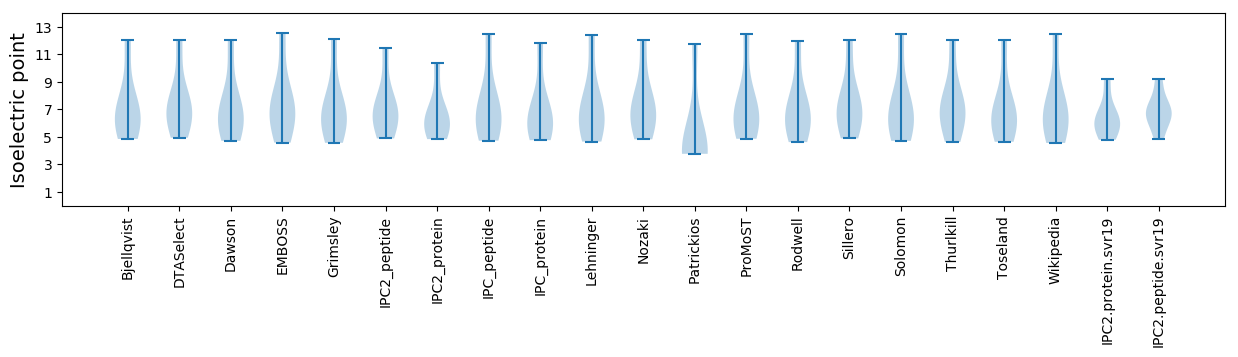
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S7J030|A0A1S7J030_9POLY Minor capsid protein OS=Rousettus aegyptiacus polyomavirus 1 OX=1904411 PE=3 SV=1
MM1 pKa = 7.25GHH3 pKa = 6.05QNGINGGLTLMTCSAMNIFLQMKK26 pKa = 9.61RR27 pKa = 11.84NPLHH31 pKa = 6.1FRR33 pKa = 11.84RR34 pKa = 11.84EE35 pKa = 4.15NHH37 pKa = 5.76HH38 pKa = 6.36HH39 pKa = 6.08QSLHH43 pKa = 4.94KK44 pKa = 8.63QHH46 pKa = 5.93RR47 pKa = 11.84QKK49 pKa = 10.51RR50 pKa = 11.84RR51 pKa = 11.84KK52 pKa = 9.03LHH54 pKa = 5.64PPKK57 pKa = 10.37IFLKK61 pKa = 9.92TFSPSS66 pKa = 2.72
MM1 pKa = 7.25GHH3 pKa = 6.05QNGINGGLTLMTCSAMNIFLQMKK26 pKa = 9.61RR27 pKa = 11.84NPLHH31 pKa = 6.1FRR33 pKa = 11.84RR34 pKa = 11.84EE35 pKa = 4.15NHH37 pKa = 5.76HH38 pKa = 6.36HH39 pKa = 6.08QSLHH43 pKa = 4.94KK44 pKa = 8.63QHH46 pKa = 5.93RR47 pKa = 11.84QKK49 pKa = 10.51RR50 pKa = 11.84RR51 pKa = 11.84KK52 pKa = 9.03LHH54 pKa = 5.64PPKK57 pKa = 10.37IFLKK61 pKa = 9.92TFSPSS66 pKa = 2.72
Molecular weight: 7.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1806 |
66 |
652 |
301.0 |
34.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.7 ± 1.646 | 1.993 ± 0.795 |
4.817 ± 0.405 | 6.977 ± 0.555 |
4.43 ± 0.553 | 6.811 ± 0.883 |
2.658 ± 0.658 | 3.987 ± 0.525 |
6.257 ± 1.356 | 10.465 ± 0.824 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.104 ± 0.483 | 4.319 ± 0.471 |
6.035 ± 0.416 | 4.374 ± 0.358 |
4.817 ± 0.808 | 6.589 ± 0.141 |
5.759 ± 0.424 | 5.205 ± 0.771 |
1.661 ± 0.531 | 4.042 ± 0.412 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
