
Crimean-Congo hemorrhagic fever virus (strain Nigeria/IbAr10200/1970) (CCHFV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Nairoviridae; Orthonairovirus; Crimean-Congo hemorrhagic fever orthonairovirus
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
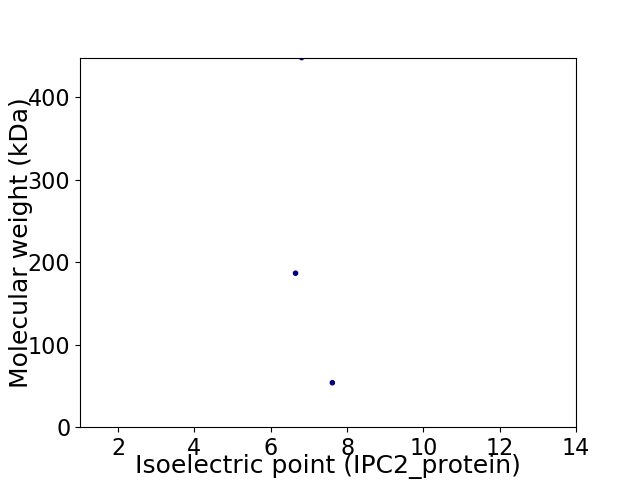
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q8JSZ3|GP_CCHFI Envelopment polyprotein OS=Crimean-Congo hemorrhagic fever virus (strain Nigeria/IbAr10200/1970) OX=652961 GN=GP PE=1 SV=1
MM1 pKa = 7.57HH2 pKa = 7.6ISLMYY7 pKa = 10.69AILCLQLCGLGEE19 pKa = 4.23THH21 pKa = 7.13GSHH24 pKa = 6.97NEE26 pKa = 3.67TRR28 pKa = 11.84HH29 pKa = 5.46NKK31 pKa = 7.96TDD33 pKa = 3.19TMTTPGDD40 pKa = 3.9NPSSEE45 pKa = 4.55PPVSTALSITLDD57 pKa = 3.78PSTVTPTTPASGLEE71 pKa = 4.25GSGEE75 pKa = 4.67VYY77 pKa = 10.3TSPPITTGSLPLSEE91 pKa = 4.77TTPEE95 pKa = 4.16LPVTTGTDD103 pKa = 3.14TLSAGDD109 pKa = 4.41VDD111 pKa = 5.53PSTQTAGGTSAPTVRR126 pKa = 11.84TSLPNSPSTPSTPQDD141 pKa = 2.88THH143 pKa = 7.73HH144 pKa = 7.12PVRR147 pKa = 11.84NLLSVTSPGPDD158 pKa = 3.06EE159 pKa = 4.34TSTPSGTGKK168 pKa = 10.28EE169 pKa = 4.31SSATSSPHH177 pKa = 5.69PVSNRR182 pKa = 11.84PPTPPATAQGPTEE195 pKa = 4.2NDD197 pKa = 2.79SHH199 pKa = 6.62NATEE203 pKa = 4.54HH204 pKa = 6.75PEE206 pKa = 4.04SLTQSATPGLMTSPTQIVHH225 pKa = 6.12PQSATPITVQDD236 pKa = 3.6THH238 pKa = 7.11PSPTNRR244 pKa = 11.84SKK246 pKa = 11.3RR247 pKa = 11.84NLKK250 pKa = 9.43MEE252 pKa = 5.4IILTLSQGLKK262 pKa = 9.96KK263 pKa = 10.84YY264 pKa = 9.74YY265 pKa = 10.85GKK267 pKa = 10.03ILRR270 pKa = 11.84LLQLTLEE277 pKa = 4.49EE278 pKa = 4.32DD279 pKa = 3.97TEE281 pKa = 5.85GLLEE285 pKa = 3.92WCKK288 pKa = 10.92RR289 pKa = 11.84NLGLDD294 pKa = 3.49CDD296 pKa = 3.88DD297 pKa = 4.45TFFQKK302 pKa = 10.48RR303 pKa = 11.84IEE305 pKa = 4.14EE306 pKa = 4.03FFITGEE312 pKa = 3.99GHH314 pKa = 6.27FNEE317 pKa = 4.43VLQFRR322 pKa = 11.84TPGTLSTTEE331 pKa = 4.03STPAGLPTAEE341 pKa = 4.2PFKK344 pKa = 11.25SYY346 pKa = 9.76FAKK349 pKa = 10.71GFLSIDD355 pKa = 3.11SGYY358 pKa = 11.25YY359 pKa = 9.14SAKK362 pKa = 10.11CYY364 pKa = 10.69SGTSNSGLQLINITRR379 pKa = 11.84HH380 pKa = 3.69STRR383 pKa = 11.84IVDD386 pKa = 3.78TPGPKK391 pKa = 8.36ITNLKK396 pKa = 8.95TINCINLKK404 pKa = 10.51ASIFKK409 pKa = 8.73EE410 pKa = 3.8HH411 pKa = 7.17RR412 pKa = 11.84EE413 pKa = 3.9VEE415 pKa = 4.36INVLLPQVAVNLSNCHH431 pKa = 4.8VVIKK435 pKa = 10.33SHH437 pKa = 5.66VCDD440 pKa = 3.72YY441 pKa = 11.57SLDD444 pKa = 3.28IDD446 pKa = 4.13GAVRR450 pKa = 11.84LPHH453 pKa = 6.95IYY455 pKa = 10.2HH456 pKa = 6.86EE457 pKa = 5.01GVFIPGTYY465 pKa = 9.93KK466 pKa = 10.32IVIDD470 pKa = 4.78KK471 pKa = 10.73KK472 pKa = 10.58NKK474 pKa = 10.24LNDD477 pKa = 3.38RR478 pKa = 11.84CTLFTDD484 pKa = 4.46CVIKK488 pKa = 10.68GRR490 pKa = 11.84EE491 pKa = 3.89VRR493 pKa = 11.84KK494 pKa = 9.13GQSVLRR500 pKa = 11.84QYY502 pKa = 9.37KK503 pKa = 8.38TEE505 pKa = 3.6IRR507 pKa = 11.84IGKK510 pKa = 9.66ASTGSRR516 pKa = 11.84RR517 pKa = 11.84LLSEE521 pKa = 4.6EE522 pKa = 4.67PSDD525 pKa = 4.1DD526 pKa = 4.31CISRR530 pKa = 11.84TQLLRR535 pKa = 11.84TEE537 pKa = 4.14TAEE540 pKa = 3.81IHH542 pKa = 6.1GDD544 pKa = 3.43NYY546 pKa = 10.95GGPGDD551 pKa = 5.55KK552 pKa = 9.4ITICNGSTIVDD563 pKa = 3.39QRR565 pKa = 11.84LGSEE569 pKa = 4.37LGCYY573 pKa = 8.48TINRR577 pKa = 11.84VRR579 pKa = 11.84SFKK582 pKa = 10.74LCEE585 pKa = 3.7NSATGKK591 pKa = 9.8NCEE594 pKa = 4.05IDD596 pKa = 3.91SVPVKK601 pKa = 10.42CRR603 pKa = 11.84QGYY606 pKa = 7.83CLRR609 pKa = 11.84ITQEE613 pKa = 3.7GRR615 pKa = 11.84GHH617 pKa = 5.81VKK619 pKa = 10.52LSRR622 pKa = 11.84GSEE625 pKa = 4.26VVLDD629 pKa = 4.32ACDD632 pKa = 3.66TSCEE636 pKa = 3.72IMIPKK641 pKa = 8.49GTGDD645 pKa = 3.54ILVDD649 pKa = 3.59CSGGQQHH656 pKa = 6.25FLKK659 pKa = 10.96DD660 pKa = 3.45NLIDD664 pKa = 4.26LGCPKK669 pKa = 10.17IPLLGKK675 pKa = 8.68MAIYY679 pKa = 9.27ICRR682 pKa = 11.84MSNHH686 pKa = 7.17PKK688 pKa = 7.76TTMAFLFWFSFGYY701 pKa = 10.71VITCILCKK709 pKa = 10.42AIFYY713 pKa = 10.41LLIIVGTLGKK723 pKa = 10.15RR724 pKa = 11.84LKK726 pKa = 10.24QYY728 pKa = 10.99RR729 pKa = 11.84EE730 pKa = 4.51LKK732 pKa = 10.09PQTCTICEE740 pKa = 4.31TTPVNAIDD748 pKa = 5.8AEE750 pKa = 4.33MHH752 pKa = 6.94DD753 pKa = 5.16LNCSYY758 pKa = 10.8NICPYY763 pKa = 9.89CASRR767 pKa = 11.84LTSDD771 pKa = 3.22GLARR775 pKa = 11.84HH776 pKa = 6.54VIQCPKK782 pKa = 10.08RR783 pKa = 11.84KK784 pKa = 9.5EE785 pKa = 4.17KK786 pKa = 11.17VEE788 pKa = 3.94EE789 pKa = 4.14TEE791 pKa = 5.09LYY793 pKa = 10.93LNLEE797 pKa = 4.78RR798 pKa = 11.84IPWVVRR804 pKa = 11.84KK805 pKa = 9.46LLQVSEE811 pKa = 4.48STGVALKK818 pKa = 10.37RR819 pKa = 11.84SSWLIVLLVLFTVSLSPVQSAPIGQGKK846 pKa = 7.84TIEE849 pKa = 4.25AYY851 pKa = 9.9RR852 pKa = 11.84ARR854 pKa = 11.84EE855 pKa = 4.61GYY857 pKa = 9.05TSICLFVLGSILFIVSCLMKK877 pKa = 10.83GLVDD881 pKa = 3.72SVGNSFFPGLSICKK895 pKa = 8.08TCSISSINGFEE906 pKa = 4.15IEE908 pKa = 4.26SHH910 pKa = 5.99KK911 pKa = 10.76CYY913 pKa = 10.36CSLFCCPYY921 pKa = 10.1CRR923 pKa = 11.84HH924 pKa = 6.54CSTDD928 pKa = 3.5KK929 pKa = 10.8EE930 pKa = 4.17IHH932 pKa = 6.39KK933 pKa = 10.28LHH935 pKa = 7.1LSICKK940 pKa = 9.65KK941 pKa = 9.91RR942 pKa = 11.84KK943 pKa = 9.05KK944 pKa = 10.38GSNVMLAVCKK954 pKa = 10.15LMCFRR959 pKa = 11.84ATMEE963 pKa = 3.88VSNRR967 pKa = 11.84ALFIRR972 pKa = 11.84SIINTTFVLCILILAVCVVSTSAVEE997 pKa = 4.21MEE999 pKa = 4.55NLPAGTWEE1007 pKa = 4.25RR1008 pKa = 11.84EE1009 pKa = 3.44EE1010 pKa = 5.16DD1011 pKa = 3.49LTNFCHH1017 pKa = 6.99QEE1019 pKa = 4.06CQVTEE1024 pKa = 4.32TEE1026 pKa = 4.39CLCPYY1031 pKa = 9.2EE1032 pKa = 4.69ALVLRR1037 pKa = 11.84KK1038 pKa = 9.11PLFLDD1043 pKa = 3.65STAKK1047 pKa = 10.34GMKK1050 pKa = 9.79NLLNSTSLEE1059 pKa = 4.0TSLSIEE1065 pKa = 4.44APWGAINVQSTYY1077 pKa = 10.95KK1078 pKa = 9.46PTVSTANIALSWSSVEE1094 pKa = 3.85HH1095 pKa = 6.69RR1096 pKa = 11.84GNKK1099 pKa = 9.27ILVSGRR1105 pKa = 11.84SEE1107 pKa = 4.73SIMKK1111 pKa = 10.41LEE1113 pKa = 3.97EE1114 pKa = 3.85RR1115 pKa = 11.84TGISWDD1121 pKa = 3.84LGVEE1125 pKa = 4.13DD1126 pKa = 4.85ASEE1129 pKa = 4.31SKK1131 pKa = 10.9LLTVSVMDD1139 pKa = 4.3LSQMYY1144 pKa = 10.35SPVFEE1149 pKa = 4.36YY1150 pKa = 10.94LSGDD1154 pKa = 3.66RR1155 pKa = 11.84QVGEE1159 pKa = 4.19WPKK1162 pKa = 9.96ATCTGDD1168 pKa = 3.32CPEE1171 pKa = 4.51RR1172 pKa = 11.84CGCTSSTCLHH1182 pKa = 6.53KK1183 pKa = 10.5EE1184 pKa = 3.88WPHH1187 pKa = 5.34SRR1189 pKa = 11.84NWRR1192 pKa = 11.84CNPTWCWGVGTGCTCCGLDD1211 pKa = 4.15VKK1213 pKa = 11.17DD1214 pKa = 5.11LFTDD1218 pKa = 3.48YY1219 pKa = 10.87MFVKK1223 pKa = 10.23WKK1225 pKa = 10.39VEE1227 pKa = 4.15YY1228 pKa = 10.15IKK1230 pKa = 10.12TEE1232 pKa = 4.4AIVCVEE1238 pKa = 4.09LTSQEE1243 pKa = 4.24RR1244 pKa = 11.84QCSLIEE1250 pKa = 4.15AGTRR1254 pKa = 11.84FNLGPVTITLSEE1266 pKa = 4.08PRR1268 pKa = 11.84NIQQKK1273 pKa = 10.1LPPEE1277 pKa = 5.18IITLHH1282 pKa = 5.77PRR1284 pKa = 11.84IEE1286 pKa = 4.17EE1287 pKa = 4.26GFFDD1291 pKa = 4.31LMHH1294 pKa = 5.39VQKK1297 pKa = 10.66VLSASTVCKK1306 pKa = 10.29LQSCTHH1312 pKa = 6.41GVPGDD1317 pKa = 3.56LQVYY1321 pKa = 10.12HH1322 pKa = 7.16IGNLLKK1328 pKa = 10.12GDD1330 pKa = 3.99KK1331 pKa = 11.03VNGHH1335 pKa = 7.8LIHH1338 pKa = 7.27KK1339 pKa = 9.78IEE1341 pKa = 4.02PHH1343 pKa = 6.59FNTSWMSWDD1352 pKa = 3.69GCDD1355 pKa = 4.67LDD1357 pKa = 5.18YY1358 pKa = 10.77YY1359 pKa = 11.18CNMGDD1364 pKa = 3.9WPSCTYY1370 pKa = 10.54TGVTQHH1376 pKa = 5.65NHH1378 pKa = 4.59ASFVNLLNIEE1388 pKa = 3.91TDD1390 pKa = 3.53YY1391 pKa = 11.39TKK1393 pKa = 10.99NFHH1396 pKa = 5.91FHH1398 pKa = 6.05SKK1400 pKa = 9.82RR1401 pKa = 11.84VTAHH1405 pKa = 6.96GDD1407 pKa = 3.46TPQLDD1412 pKa = 3.89LKK1414 pKa = 11.05ARR1416 pKa = 11.84PTYY1419 pKa = 10.09GAGEE1423 pKa = 3.97ITVLVEE1429 pKa = 3.85VADD1432 pKa = 4.23MEE1434 pKa = 4.57LHH1436 pKa = 5.51TKK1438 pKa = 10.14KK1439 pKa = 10.37IEE1441 pKa = 3.75ISGLKK1446 pKa = 9.42FASLACTGCYY1456 pKa = 9.88ACSSGISCKK1465 pKa = 10.1VRR1467 pKa = 11.84IHH1469 pKa = 7.01VDD1471 pKa = 3.27EE1472 pKa = 5.16PDD1474 pKa = 3.3EE1475 pKa = 4.12LTVHH1479 pKa = 6.18VKK1481 pKa = 10.51SDD1483 pKa = 3.8DD1484 pKa = 3.91PDD1486 pKa = 3.58VVAASSSLMARR1497 pKa = 11.84KK1498 pKa = 9.96LEE1500 pKa = 4.28FGTDD1504 pKa = 3.28STFKK1508 pKa = 10.78AFSAMPKK1515 pKa = 9.55TSLCFYY1521 pKa = 9.63IVEE1524 pKa = 4.26RR1525 pKa = 11.84EE1526 pKa = 4.14HH1527 pKa = 7.14CKK1529 pKa = 10.45SCSEE1533 pKa = 4.19EE1534 pKa = 4.02DD1535 pKa = 3.25TKK1537 pKa = 11.58KK1538 pKa = 10.76CVNTKK1543 pKa = 10.45LEE1545 pKa = 4.13QPQSILIEE1553 pKa = 4.15HH1554 pKa = 7.26KK1555 pKa = 9.71GTIIGKK1561 pKa = 8.99QNSTCTAKK1569 pKa = 10.64ASCWLEE1575 pKa = 3.93SVKK1578 pKa = 11.0SFFYY1582 pKa = 10.73GLKK1585 pKa = 10.61NMLSGIFGNVFMGIFLFLAPFILLILFFMFGWRR1618 pKa = 11.84ILFCFKK1624 pKa = 10.05CCRR1627 pKa = 11.84RR1628 pKa = 11.84TRR1630 pKa = 11.84GLFKK1634 pKa = 10.65YY1635 pKa = 10.11RR1636 pKa = 11.84HH1637 pKa = 6.01LKK1639 pKa = 10.46DD1640 pKa = 4.44DD1641 pKa = 3.72EE1642 pKa = 4.22EE1643 pKa = 4.17TGYY1646 pKa = 10.9RR1647 pKa = 11.84RR1648 pKa = 11.84IIEE1651 pKa = 4.03KK1652 pKa = 10.75LNNKK1656 pKa = 8.89KK1657 pKa = 10.48GKK1659 pKa = 9.99NKK1661 pKa = 10.26LLDD1664 pKa = 3.64GEE1666 pKa = 4.44RR1667 pKa = 11.84LADD1670 pKa = 3.78RR1671 pKa = 11.84RR1672 pKa = 11.84IAEE1675 pKa = 4.21LFSTKK1680 pKa = 7.31THH1682 pKa = 6.46IGG1684 pKa = 3.3
MM1 pKa = 7.57HH2 pKa = 7.6ISLMYY7 pKa = 10.69AILCLQLCGLGEE19 pKa = 4.23THH21 pKa = 7.13GSHH24 pKa = 6.97NEE26 pKa = 3.67TRR28 pKa = 11.84HH29 pKa = 5.46NKK31 pKa = 7.96TDD33 pKa = 3.19TMTTPGDD40 pKa = 3.9NPSSEE45 pKa = 4.55PPVSTALSITLDD57 pKa = 3.78PSTVTPTTPASGLEE71 pKa = 4.25GSGEE75 pKa = 4.67VYY77 pKa = 10.3TSPPITTGSLPLSEE91 pKa = 4.77TTPEE95 pKa = 4.16LPVTTGTDD103 pKa = 3.14TLSAGDD109 pKa = 4.41VDD111 pKa = 5.53PSTQTAGGTSAPTVRR126 pKa = 11.84TSLPNSPSTPSTPQDD141 pKa = 2.88THH143 pKa = 7.73HH144 pKa = 7.12PVRR147 pKa = 11.84NLLSVTSPGPDD158 pKa = 3.06EE159 pKa = 4.34TSTPSGTGKK168 pKa = 10.28EE169 pKa = 4.31SSATSSPHH177 pKa = 5.69PVSNRR182 pKa = 11.84PPTPPATAQGPTEE195 pKa = 4.2NDD197 pKa = 2.79SHH199 pKa = 6.62NATEE203 pKa = 4.54HH204 pKa = 6.75PEE206 pKa = 4.04SLTQSATPGLMTSPTQIVHH225 pKa = 6.12PQSATPITVQDD236 pKa = 3.6THH238 pKa = 7.11PSPTNRR244 pKa = 11.84SKK246 pKa = 11.3RR247 pKa = 11.84NLKK250 pKa = 9.43MEE252 pKa = 5.4IILTLSQGLKK262 pKa = 9.96KK263 pKa = 10.84YY264 pKa = 9.74YY265 pKa = 10.85GKK267 pKa = 10.03ILRR270 pKa = 11.84LLQLTLEE277 pKa = 4.49EE278 pKa = 4.32DD279 pKa = 3.97TEE281 pKa = 5.85GLLEE285 pKa = 3.92WCKK288 pKa = 10.92RR289 pKa = 11.84NLGLDD294 pKa = 3.49CDD296 pKa = 3.88DD297 pKa = 4.45TFFQKK302 pKa = 10.48RR303 pKa = 11.84IEE305 pKa = 4.14EE306 pKa = 4.03FFITGEE312 pKa = 3.99GHH314 pKa = 6.27FNEE317 pKa = 4.43VLQFRR322 pKa = 11.84TPGTLSTTEE331 pKa = 4.03STPAGLPTAEE341 pKa = 4.2PFKK344 pKa = 11.25SYY346 pKa = 9.76FAKK349 pKa = 10.71GFLSIDD355 pKa = 3.11SGYY358 pKa = 11.25YY359 pKa = 9.14SAKK362 pKa = 10.11CYY364 pKa = 10.69SGTSNSGLQLINITRR379 pKa = 11.84HH380 pKa = 3.69STRR383 pKa = 11.84IVDD386 pKa = 3.78TPGPKK391 pKa = 8.36ITNLKK396 pKa = 8.95TINCINLKK404 pKa = 10.51ASIFKK409 pKa = 8.73EE410 pKa = 3.8HH411 pKa = 7.17RR412 pKa = 11.84EE413 pKa = 3.9VEE415 pKa = 4.36INVLLPQVAVNLSNCHH431 pKa = 4.8VVIKK435 pKa = 10.33SHH437 pKa = 5.66VCDD440 pKa = 3.72YY441 pKa = 11.57SLDD444 pKa = 3.28IDD446 pKa = 4.13GAVRR450 pKa = 11.84LPHH453 pKa = 6.95IYY455 pKa = 10.2HH456 pKa = 6.86EE457 pKa = 5.01GVFIPGTYY465 pKa = 9.93KK466 pKa = 10.32IVIDD470 pKa = 4.78KK471 pKa = 10.73KK472 pKa = 10.58NKK474 pKa = 10.24LNDD477 pKa = 3.38RR478 pKa = 11.84CTLFTDD484 pKa = 4.46CVIKK488 pKa = 10.68GRR490 pKa = 11.84EE491 pKa = 3.89VRR493 pKa = 11.84KK494 pKa = 9.13GQSVLRR500 pKa = 11.84QYY502 pKa = 9.37KK503 pKa = 8.38TEE505 pKa = 3.6IRR507 pKa = 11.84IGKK510 pKa = 9.66ASTGSRR516 pKa = 11.84RR517 pKa = 11.84LLSEE521 pKa = 4.6EE522 pKa = 4.67PSDD525 pKa = 4.1DD526 pKa = 4.31CISRR530 pKa = 11.84TQLLRR535 pKa = 11.84TEE537 pKa = 4.14TAEE540 pKa = 3.81IHH542 pKa = 6.1GDD544 pKa = 3.43NYY546 pKa = 10.95GGPGDD551 pKa = 5.55KK552 pKa = 9.4ITICNGSTIVDD563 pKa = 3.39QRR565 pKa = 11.84LGSEE569 pKa = 4.37LGCYY573 pKa = 8.48TINRR577 pKa = 11.84VRR579 pKa = 11.84SFKK582 pKa = 10.74LCEE585 pKa = 3.7NSATGKK591 pKa = 9.8NCEE594 pKa = 4.05IDD596 pKa = 3.91SVPVKK601 pKa = 10.42CRR603 pKa = 11.84QGYY606 pKa = 7.83CLRR609 pKa = 11.84ITQEE613 pKa = 3.7GRR615 pKa = 11.84GHH617 pKa = 5.81VKK619 pKa = 10.52LSRR622 pKa = 11.84GSEE625 pKa = 4.26VVLDD629 pKa = 4.32ACDD632 pKa = 3.66TSCEE636 pKa = 3.72IMIPKK641 pKa = 8.49GTGDD645 pKa = 3.54ILVDD649 pKa = 3.59CSGGQQHH656 pKa = 6.25FLKK659 pKa = 10.96DD660 pKa = 3.45NLIDD664 pKa = 4.26LGCPKK669 pKa = 10.17IPLLGKK675 pKa = 8.68MAIYY679 pKa = 9.27ICRR682 pKa = 11.84MSNHH686 pKa = 7.17PKK688 pKa = 7.76TTMAFLFWFSFGYY701 pKa = 10.71VITCILCKK709 pKa = 10.42AIFYY713 pKa = 10.41LLIIVGTLGKK723 pKa = 10.15RR724 pKa = 11.84LKK726 pKa = 10.24QYY728 pKa = 10.99RR729 pKa = 11.84EE730 pKa = 4.51LKK732 pKa = 10.09PQTCTICEE740 pKa = 4.31TTPVNAIDD748 pKa = 5.8AEE750 pKa = 4.33MHH752 pKa = 6.94DD753 pKa = 5.16LNCSYY758 pKa = 10.8NICPYY763 pKa = 9.89CASRR767 pKa = 11.84LTSDD771 pKa = 3.22GLARR775 pKa = 11.84HH776 pKa = 6.54VIQCPKK782 pKa = 10.08RR783 pKa = 11.84KK784 pKa = 9.5EE785 pKa = 4.17KK786 pKa = 11.17VEE788 pKa = 3.94EE789 pKa = 4.14TEE791 pKa = 5.09LYY793 pKa = 10.93LNLEE797 pKa = 4.78RR798 pKa = 11.84IPWVVRR804 pKa = 11.84KK805 pKa = 9.46LLQVSEE811 pKa = 4.48STGVALKK818 pKa = 10.37RR819 pKa = 11.84SSWLIVLLVLFTVSLSPVQSAPIGQGKK846 pKa = 7.84TIEE849 pKa = 4.25AYY851 pKa = 9.9RR852 pKa = 11.84ARR854 pKa = 11.84EE855 pKa = 4.61GYY857 pKa = 9.05TSICLFVLGSILFIVSCLMKK877 pKa = 10.83GLVDD881 pKa = 3.72SVGNSFFPGLSICKK895 pKa = 8.08TCSISSINGFEE906 pKa = 4.15IEE908 pKa = 4.26SHH910 pKa = 5.99KK911 pKa = 10.76CYY913 pKa = 10.36CSLFCCPYY921 pKa = 10.1CRR923 pKa = 11.84HH924 pKa = 6.54CSTDD928 pKa = 3.5KK929 pKa = 10.8EE930 pKa = 4.17IHH932 pKa = 6.39KK933 pKa = 10.28LHH935 pKa = 7.1LSICKK940 pKa = 9.65KK941 pKa = 9.91RR942 pKa = 11.84KK943 pKa = 9.05KK944 pKa = 10.38GSNVMLAVCKK954 pKa = 10.15LMCFRR959 pKa = 11.84ATMEE963 pKa = 3.88VSNRR967 pKa = 11.84ALFIRR972 pKa = 11.84SIINTTFVLCILILAVCVVSTSAVEE997 pKa = 4.21MEE999 pKa = 4.55NLPAGTWEE1007 pKa = 4.25RR1008 pKa = 11.84EE1009 pKa = 3.44EE1010 pKa = 5.16DD1011 pKa = 3.49LTNFCHH1017 pKa = 6.99QEE1019 pKa = 4.06CQVTEE1024 pKa = 4.32TEE1026 pKa = 4.39CLCPYY1031 pKa = 9.2EE1032 pKa = 4.69ALVLRR1037 pKa = 11.84KK1038 pKa = 9.11PLFLDD1043 pKa = 3.65STAKK1047 pKa = 10.34GMKK1050 pKa = 9.79NLLNSTSLEE1059 pKa = 4.0TSLSIEE1065 pKa = 4.44APWGAINVQSTYY1077 pKa = 10.95KK1078 pKa = 9.46PTVSTANIALSWSSVEE1094 pKa = 3.85HH1095 pKa = 6.69RR1096 pKa = 11.84GNKK1099 pKa = 9.27ILVSGRR1105 pKa = 11.84SEE1107 pKa = 4.73SIMKK1111 pKa = 10.41LEE1113 pKa = 3.97EE1114 pKa = 3.85RR1115 pKa = 11.84TGISWDD1121 pKa = 3.84LGVEE1125 pKa = 4.13DD1126 pKa = 4.85ASEE1129 pKa = 4.31SKK1131 pKa = 10.9LLTVSVMDD1139 pKa = 4.3LSQMYY1144 pKa = 10.35SPVFEE1149 pKa = 4.36YY1150 pKa = 10.94LSGDD1154 pKa = 3.66RR1155 pKa = 11.84QVGEE1159 pKa = 4.19WPKK1162 pKa = 9.96ATCTGDD1168 pKa = 3.32CPEE1171 pKa = 4.51RR1172 pKa = 11.84CGCTSSTCLHH1182 pKa = 6.53KK1183 pKa = 10.5EE1184 pKa = 3.88WPHH1187 pKa = 5.34SRR1189 pKa = 11.84NWRR1192 pKa = 11.84CNPTWCWGVGTGCTCCGLDD1211 pKa = 4.15VKK1213 pKa = 11.17DD1214 pKa = 5.11LFTDD1218 pKa = 3.48YY1219 pKa = 10.87MFVKK1223 pKa = 10.23WKK1225 pKa = 10.39VEE1227 pKa = 4.15YY1228 pKa = 10.15IKK1230 pKa = 10.12TEE1232 pKa = 4.4AIVCVEE1238 pKa = 4.09LTSQEE1243 pKa = 4.24RR1244 pKa = 11.84QCSLIEE1250 pKa = 4.15AGTRR1254 pKa = 11.84FNLGPVTITLSEE1266 pKa = 4.08PRR1268 pKa = 11.84NIQQKK1273 pKa = 10.1LPPEE1277 pKa = 5.18IITLHH1282 pKa = 5.77PRR1284 pKa = 11.84IEE1286 pKa = 4.17EE1287 pKa = 4.26GFFDD1291 pKa = 4.31LMHH1294 pKa = 5.39VQKK1297 pKa = 10.66VLSASTVCKK1306 pKa = 10.29LQSCTHH1312 pKa = 6.41GVPGDD1317 pKa = 3.56LQVYY1321 pKa = 10.12HH1322 pKa = 7.16IGNLLKK1328 pKa = 10.12GDD1330 pKa = 3.99KK1331 pKa = 11.03VNGHH1335 pKa = 7.8LIHH1338 pKa = 7.27KK1339 pKa = 9.78IEE1341 pKa = 4.02PHH1343 pKa = 6.59FNTSWMSWDD1352 pKa = 3.69GCDD1355 pKa = 4.67LDD1357 pKa = 5.18YY1358 pKa = 10.77YY1359 pKa = 11.18CNMGDD1364 pKa = 3.9WPSCTYY1370 pKa = 10.54TGVTQHH1376 pKa = 5.65NHH1378 pKa = 4.59ASFVNLLNIEE1388 pKa = 3.91TDD1390 pKa = 3.53YY1391 pKa = 11.39TKK1393 pKa = 10.99NFHH1396 pKa = 5.91FHH1398 pKa = 6.05SKK1400 pKa = 9.82RR1401 pKa = 11.84VTAHH1405 pKa = 6.96GDD1407 pKa = 3.46TPQLDD1412 pKa = 3.89LKK1414 pKa = 11.05ARR1416 pKa = 11.84PTYY1419 pKa = 10.09GAGEE1423 pKa = 3.97ITVLVEE1429 pKa = 3.85VADD1432 pKa = 4.23MEE1434 pKa = 4.57LHH1436 pKa = 5.51TKK1438 pKa = 10.14KK1439 pKa = 10.37IEE1441 pKa = 3.75ISGLKK1446 pKa = 9.42FASLACTGCYY1456 pKa = 9.88ACSSGISCKK1465 pKa = 10.1VRR1467 pKa = 11.84IHH1469 pKa = 7.01VDD1471 pKa = 3.27EE1472 pKa = 5.16PDD1474 pKa = 3.3EE1475 pKa = 4.12LTVHH1479 pKa = 6.18VKK1481 pKa = 10.51SDD1483 pKa = 3.8DD1484 pKa = 3.91PDD1486 pKa = 3.58VVAASSSLMARR1497 pKa = 11.84KK1498 pKa = 9.96LEE1500 pKa = 4.28FGTDD1504 pKa = 3.28STFKK1508 pKa = 10.78AFSAMPKK1515 pKa = 9.55TSLCFYY1521 pKa = 9.63IVEE1524 pKa = 4.26RR1525 pKa = 11.84EE1526 pKa = 4.14HH1527 pKa = 7.14CKK1529 pKa = 10.45SCSEE1533 pKa = 4.19EE1534 pKa = 4.02DD1535 pKa = 3.25TKK1537 pKa = 11.58KK1538 pKa = 10.76CVNTKK1543 pKa = 10.45LEE1545 pKa = 4.13QPQSILIEE1553 pKa = 4.15HH1554 pKa = 7.26KK1555 pKa = 9.71GTIIGKK1561 pKa = 8.99QNSTCTAKK1569 pKa = 10.64ASCWLEE1575 pKa = 3.93SVKK1578 pKa = 11.0SFFYY1582 pKa = 10.73GLKK1585 pKa = 10.61NMLSGIFGNVFMGIFLFLAPFILLILFFMFGWRR1618 pKa = 11.84ILFCFKK1624 pKa = 10.05CCRR1627 pKa = 11.84RR1628 pKa = 11.84TRR1630 pKa = 11.84GLFKK1634 pKa = 10.65YY1635 pKa = 10.11RR1636 pKa = 11.84HH1637 pKa = 6.01LKK1639 pKa = 10.46DD1640 pKa = 4.44DD1641 pKa = 3.72EE1642 pKa = 4.22EE1643 pKa = 4.17TGYY1646 pKa = 10.9RR1647 pKa = 11.84RR1648 pKa = 11.84IIEE1651 pKa = 4.03KK1652 pKa = 10.75LNNKK1656 pKa = 8.89KK1657 pKa = 10.48GKK1659 pKa = 9.99NKK1661 pKa = 10.26LLDD1664 pKa = 3.64GEE1666 pKa = 4.44RR1667 pKa = 11.84LADD1670 pKa = 3.78RR1671 pKa = 11.84RR1672 pKa = 11.84IAEE1675 pKa = 4.21LFSTKK1680 pKa = 7.31THH1682 pKa = 6.46IGG1684 pKa = 3.3
Molecular weight: 186.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q6TQR6|L_CCHFI RNA-directed RNA polymerase L OS=Crimean-Congo hemorrhagic fever virus (strain Nigeria/IbAr10200/1970) OX=652961 GN=L PE=1 SV=2
MM1 pKa = 7.37EE2 pKa = 5.21NKK4 pKa = 9.92IEE6 pKa = 4.12VNNKK10 pKa = 10.24DD11 pKa = 3.26EE12 pKa = 4.19MNRR15 pKa = 11.84WFEE18 pKa = 4.04EE19 pKa = 4.28FKK21 pKa = 10.89KK22 pKa = 11.23GNGLVDD28 pKa = 4.02TFTNSYY34 pKa = 9.74SFCEE38 pKa = 4.17SVPNLDD44 pKa = 3.08RR45 pKa = 11.84FVFQMASATDD55 pKa = 3.9DD56 pKa = 3.63AQKK59 pKa = 11.29DD60 pKa = 3.88SIYY63 pKa = 11.16ASALVEE69 pKa = 4.03ATKK72 pKa = 10.54FCAPIYY78 pKa = 8.27EE79 pKa = 4.62CAWVSSTGIVKK90 pKa = 10.41KK91 pKa = 10.38GLEE94 pKa = 4.12WFEE97 pKa = 4.62KK98 pKa = 10.21NAGTIKK104 pKa = 10.31SWDD107 pKa = 3.34EE108 pKa = 4.2SYY110 pKa = 10.95TEE112 pKa = 4.62LKK114 pKa = 10.66VDD116 pKa = 3.58VPKK119 pKa = 10.47IEE121 pKa = 4.08QLTGYY126 pKa = 9.0QQAALKK132 pKa = 8.35WRR134 pKa = 11.84KK135 pKa = 10.11DD136 pKa = 2.81IGFRR140 pKa = 11.84VNANTAALSNKK151 pKa = 8.73VLAEE155 pKa = 3.85YY156 pKa = 9.94KK157 pKa = 10.43VPGEE161 pKa = 3.7IVMSVKK167 pKa = 10.47EE168 pKa = 4.32MLSDD172 pKa = 3.71MIRR175 pKa = 11.84RR176 pKa = 11.84RR177 pKa = 11.84NLILNRR183 pKa = 11.84GGDD186 pKa = 3.89EE187 pKa = 4.11NPRR190 pKa = 11.84GPVSHH195 pKa = 6.54EE196 pKa = 3.72HH197 pKa = 6.3VDD199 pKa = 3.04WCRR202 pKa = 11.84EE203 pKa = 3.93FVKK206 pKa = 10.55GKK208 pKa = 10.74YY209 pKa = 9.71IMAFNPPWGDD219 pKa = 2.94INKK222 pKa = 9.96SGRR225 pKa = 11.84SGIALVATGLAKK237 pKa = 10.52LAEE240 pKa = 4.36TEE242 pKa = 4.17GKK244 pKa = 10.88GIFDD248 pKa = 3.72EE249 pKa = 4.49AKK251 pKa = 9.15KK252 pKa = 8.93TVEE255 pKa = 4.14ALNGYY260 pKa = 8.91LDD262 pKa = 3.51KK263 pKa = 11.45HH264 pKa = 7.28KK265 pKa = 11.23DD266 pKa = 3.49EE267 pKa = 4.59VDD269 pKa = 3.27RR270 pKa = 11.84ASADD274 pKa = 3.51SMITNLLKK282 pKa = 10.7HH283 pKa = 5.78IAKK286 pKa = 10.09AQEE289 pKa = 4.04LYY291 pKa = 10.76KK292 pKa = 10.97NSSALRR298 pKa = 11.84AQSAQIDD305 pKa = 4.09TAFSSYY311 pKa = 9.79YY312 pKa = 8.44WLYY315 pKa = 10.2KK316 pKa = 10.7AGVTPEE322 pKa = 4.05TFPTVSQFLFEE333 pKa = 5.15LGKK336 pKa = 10.1QPRR339 pKa = 11.84GTKK342 pKa = 9.88KK343 pKa = 9.34MKK345 pKa = 10.11KK346 pKa = 9.65ALLSTPMKK354 pKa = 9.37WGKK357 pKa = 10.08KK358 pKa = 9.13LYY360 pKa = 10.41EE361 pKa = 4.35LFADD365 pKa = 5.93DD366 pKa = 5.64SFQQNRR372 pKa = 11.84IYY374 pKa = 9.43MHH376 pKa = 6.84PAVLTAGRR384 pKa = 11.84ISEE387 pKa = 4.14MGVCFGTIPVANPDD401 pKa = 3.76DD402 pKa = 4.06AAQGSGHH409 pKa = 6.01TKK411 pKa = 10.35SILNLRR417 pKa = 11.84TNTEE421 pKa = 4.21TNNPCAKK428 pKa = 9.85TIVKK432 pKa = 10.01LFEE435 pKa = 4.02VQKK438 pKa = 9.89TGFNIQDD445 pKa = 3.36MDD447 pKa = 3.69IVASEE452 pKa = 4.34HH453 pKa = 6.72LLHH456 pKa = 6.59QSLVGKK462 pKa = 9.72QSPFQNAYY470 pKa = 8.7NVKK473 pKa = 10.46GNATSANIII482 pKa = 3.84
MM1 pKa = 7.37EE2 pKa = 5.21NKK4 pKa = 9.92IEE6 pKa = 4.12VNNKK10 pKa = 10.24DD11 pKa = 3.26EE12 pKa = 4.19MNRR15 pKa = 11.84WFEE18 pKa = 4.04EE19 pKa = 4.28FKK21 pKa = 10.89KK22 pKa = 11.23GNGLVDD28 pKa = 4.02TFTNSYY34 pKa = 9.74SFCEE38 pKa = 4.17SVPNLDD44 pKa = 3.08RR45 pKa = 11.84FVFQMASATDD55 pKa = 3.9DD56 pKa = 3.63AQKK59 pKa = 11.29DD60 pKa = 3.88SIYY63 pKa = 11.16ASALVEE69 pKa = 4.03ATKK72 pKa = 10.54FCAPIYY78 pKa = 8.27EE79 pKa = 4.62CAWVSSTGIVKK90 pKa = 10.41KK91 pKa = 10.38GLEE94 pKa = 4.12WFEE97 pKa = 4.62KK98 pKa = 10.21NAGTIKK104 pKa = 10.31SWDD107 pKa = 3.34EE108 pKa = 4.2SYY110 pKa = 10.95TEE112 pKa = 4.62LKK114 pKa = 10.66VDD116 pKa = 3.58VPKK119 pKa = 10.47IEE121 pKa = 4.08QLTGYY126 pKa = 9.0QQAALKK132 pKa = 8.35WRR134 pKa = 11.84KK135 pKa = 10.11DD136 pKa = 2.81IGFRR140 pKa = 11.84VNANTAALSNKK151 pKa = 8.73VLAEE155 pKa = 3.85YY156 pKa = 9.94KK157 pKa = 10.43VPGEE161 pKa = 3.7IVMSVKK167 pKa = 10.47EE168 pKa = 4.32MLSDD172 pKa = 3.71MIRR175 pKa = 11.84RR176 pKa = 11.84RR177 pKa = 11.84NLILNRR183 pKa = 11.84GGDD186 pKa = 3.89EE187 pKa = 4.11NPRR190 pKa = 11.84GPVSHH195 pKa = 6.54EE196 pKa = 3.72HH197 pKa = 6.3VDD199 pKa = 3.04WCRR202 pKa = 11.84EE203 pKa = 3.93FVKK206 pKa = 10.55GKK208 pKa = 10.74YY209 pKa = 9.71IMAFNPPWGDD219 pKa = 2.94INKK222 pKa = 9.96SGRR225 pKa = 11.84SGIALVATGLAKK237 pKa = 10.52LAEE240 pKa = 4.36TEE242 pKa = 4.17GKK244 pKa = 10.88GIFDD248 pKa = 3.72EE249 pKa = 4.49AKK251 pKa = 9.15KK252 pKa = 8.93TVEE255 pKa = 4.14ALNGYY260 pKa = 8.91LDD262 pKa = 3.51KK263 pKa = 11.45HH264 pKa = 7.28KK265 pKa = 11.23DD266 pKa = 3.49EE267 pKa = 4.59VDD269 pKa = 3.27RR270 pKa = 11.84ASADD274 pKa = 3.51SMITNLLKK282 pKa = 10.7HH283 pKa = 5.78IAKK286 pKa = 10.09AQEE289 pKa = 4.04LYY291 pKa = 10.76KK292 pKa = 10.97NSSALRR298 pKa = 11.84AQSAQIDD305 pKa = 4.09TAFSSYY311 pKa = 9.79YY312 pKa = 8.44WLYY315 pKa = 10.2KK316 pKa = 10.7AGVTPEE322 pKa = 4.05TFPTVSQFLFEE333 pKa = 5.15LGKK336 pKa = 10.1QPRR339 pKa = 11.84GTKK342 pKa = 9.88KK343 pKa = 9.34MKK345 pKa = 10.11KK346 pKa = 9.65ALLSTPMKK354 pKa = 9.37WGKK357 pKa = 10.08KK358 pKa = 9.13LYY360 pKa = 10.41EE361 pKa = 4.35LFADD365 pKa = 5.93DD366 pKa = 5.64SFQQNRR372 pKa = 11.84IYY374 pKa = 9.43MHH376 pKa = 6.84PAVLTAGRR384 pKa = 11.84ISEE387 pKa = 4.14MGVCFGTIPVANPDD401 pKa = 3.76DD402 pKa = 4.06AAQGSGHH409 pKa = 6.01TKK411 pKa = 10.35SILNLRR417 pKa = 11.84TNTEE421 pKa = 4.21TNNPCAKK428 pKa = 9.85TIVKK432 pKa = 10.01LFEE435 pKa = 4.02VQKK438 pKa = 9.89TGFNIQDD445 pKa = 3.36MDD447 pKa = 3.69IVASEE452 pKa = 4.34HH453 pKa = 6.72LLHH456 pKa = 6.59QSLVGKK462 pKa = 9.72QSPFQNAYY470 pKa = 8.7NVKK473 pKa = 10.46GNATSANIII482 pKa = 3.84
Molecular weight: 53.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6111 |
482 |
3945 |
2037.0 |
229.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.615 ± 1.122 | 2.995 ± 0.853 |
5.106 ± 0.423 | 6.824 ± 0.296 |
4.058 ± 0.249 | 5.236 ± 0.859 |
2.307 ± 0.354 | 5.989 ± 0.231 |
7.266 ± 0.664 | 11.193 ± 1.369 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.127 ± 0.221 | 4.893 ± 0.571 |
3.551 ± 0.746 | 3.42 ± 0.352 |
4.811 ± 0.429 | 9.18 ± 0.66 |
6.758 ± 1.04 | 6.055 ± 0.087 |
1.064 ± 0.222 | 2.553 ± 0.149 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
