
Spring viraemia of carp virus (strain ATCC VR-1390)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Sprivivirus; Spring viremia of carp virus
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
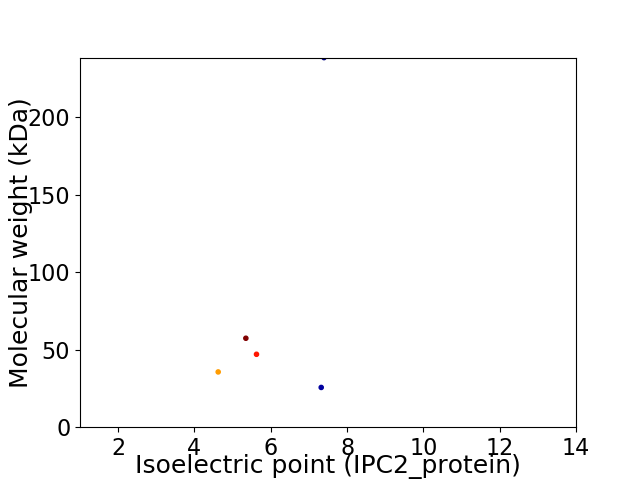
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
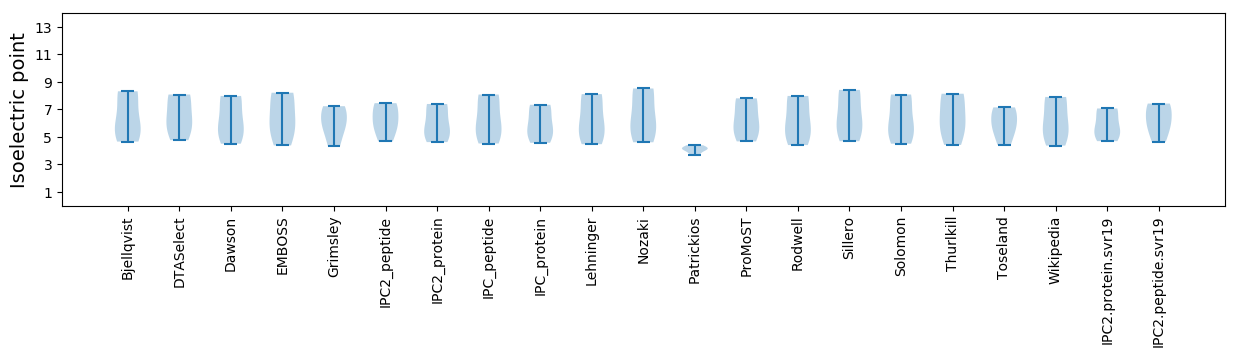
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q91DS6|Q91DS6_SVCVA Nucleocapsid protein OS=Spring viraemia of carp virus (strain ATCC VR-1390) OX=1559360 GN=N PE=4 SV=1
MM1 pKa = 7.47SLHH4 pKa = 6.48SKK6 pKa = 10.52LSEE9 pKa = 4.14SLKK12 pKa = 10.81AYY14 pKa = 10.49ADD16 pKa = 3.53LDD18 pKa = 3.84KK19 pKa = 10.69TVKK22 pKa = 10.43EE23 pKa = 4.03IEE25 pKa = 4.28EE26 pKa = 4.29QVSSMEE32 pKa = 4.0EE33 pKa = 3.94PVPKK37 pKa = 8.94TVKK40 pKa = 10.32YY41 pKa = 8.6VTFEE45 pKa = 3.94EE46 pKa = 4.52DD47 pKa = 3.0LSEE50 pKa = 4.5EE51 pKa = 4.15EE52 pKa = 4.5WEE54 pKa = 4.43SDD56 pKa = 4.86SGDD59 pKa = 3.9DD60 pKa = 5.7DD61 pKa = 4.62EE62 pKa = 7.84DD63 pKa = 6.23SIDD66 pKa = 4.52DD67 pKa = 4.25SLIPDD72 pKa = 3.93YY73 pKa = 11.01LRR75 pKa = 11.84EE76 pKa = 4.16SSSITVDD83 pKa = 3.94EE84 pKa = 5.01DD85 pKa = 4.33EE86 pKa = 4.97EE87 pKa = 4.23DD88 pKa = 3.41QKK90 pKa = 11.63EE91 pKa = 4.07DD92 pKa = 4.1RR93 pKa = 11.84EE94 pKa = 4.25EE95 pKa = 4.15HH96 pKa = 6.39LPTVSWEE103 pKa = 4.05EE104 pKa = 4.03EE105 pKa = 3.86PTGIDD110 pKa = 2.72IGFGPGIVMPSVSNHH125 pKa = 5.61EE126 pKa = 4.11GGTYY130 pKa = 9.91VRR132 pKa = 11.84YY133 pKa = 10.17NGLGGIDD140 pKa = 4.39PNCKK144 pKa = 10.08DD145 pKa = 5.09LISKK149 pKa = 7.7MMRR152 pKa = 11.84SLIGQIGNKK161 pKa = 9.32YY162 pKa = 10.51GYY164 pKa = 10.63DD165 pKa = 3.29IDD167 pKa = 5.26LFDD170 pKa = 3.78YY171 pKa = 11.0QGDD174 pKa = 3.78FLEE177 pKa = 4.56VFLPHH182 pKa = 6.85KK183 pKa = 9.94PSKK186 pKa = 10.61EE187 pKa = 3.82DD188 pKa = 3.1VRR190 pKa = 11.84PDD192 pKa = 2.86IRR194 pKa = 11.84IGKK197 pKa = 9.19KK198 pKa = 9.77NEE200 pKa = 3.82EE201 pKa = 4.3GTSKK205 pKa = 10.38QVSKK209 pKa = 10.55PRR211 pKa = 11.84EE212 pKa = 3.73KK213 pKa = 10.86EE214 pKa = 3.94KK215 pKa = 10.77IVLKK219 pKa = 10.19TGDD222 pKa = 3.28EE223 pKa = 4.34CGRR226 pKa = 11.84FPMNKK231 pKa = 8.37EE232 pKa = 3.59AKK234 pKa = 9.41KK235 pKa = 10.55RR236 pKa = 11.84EE237 pKa = 4.09PEE239 pKa = 4.18GLWEE243 pKa = 4.12VMKK246 pKa = 10.93VLSVQFDD253 pKa = 3.36PWKK256 pKa = 10.41EE257 pKa = 3.96DD258 pKa = 3.66EE259 pKa = 5.12PPLSLTIRR267 pKa = 11.84DD268 pKa = 3.82LFISEE273 pKa = 4.35SEE275 pKa = 4.08FRR277 pKa = 11.84LHH279 pKa = 6.92CNHH282 pKa = 6.13SQTEE286 pKa = 4.08RR287 pKa = 11.84EE288 pKa = 3.94MALVGIKK295 pKa = 10.07LRR297 pKa = 11.84RR298 pKa = 11.84LYY300 pKa = 11.05NKK302 pKa = 10.09LYY304 pKa = 10.24QKK306 pKa = 10.81YY307 pKa = 9.63RR308 pKa = 11.84LL309 pKa = 3.67
MM1 pKa = 7.47SLHH4 pKa = 6.48SKK6 pKa = 10.52LSEE9 pKa = 4.14SLKK12 pKa = 10.81AYY14 pKa = 10.49ADD16 pKa = 3.53LDD18 pKa = 3.84KK19 pKa = 10.69TVKK22 pKa = 10.43EE23 pKa = 4.03IEE25 pKa = 4.28EE26 pKa = 4.29QVSSMEE32 pKa = 4.0EE33 pKa = 3.94PVPKK37 pKa = 8.94TVKK40 pKa = 10.32YY41 pKa = 8.6VTFEE45 pKa = 3.94EE46 pKa = 4.52DD47 pKa = 3.0LSEE50 pKa = 4.5EE51 pKa = 4.15EE52 pKa = 4.5WEE54 pKa = 4.43SDD56 pKa = 4.86SGDD59 pKa = 3.9DD60 pKa = 5.7DD61 pKa = 4.62EE62 pKa = 7.84DD63 pKa = 6.23SIDD66 pKa = 4.52DD67 pKa = 4.25SLIPDD72 pKa = 3.93YY73 pKa = 11.01LRR75 pKa = 11.84EE76 pKa = 4.16SSSITVDD83 pKa = 3.94EE84 pKa = 5.01DD85 pKa = 4.33EE86 pKa = 4.97EE87 pKa = 4.23DD88 pKa = 3.41QKK90 pKa = 11.63EE91 pKa = 4.07DD92 pKa = 4.1RR93 pKa = 11.84EE94 pKa = 4.25EE95 pKa = 4.15HH96 pKa = 6.39LPTVSWEE103 pKa = 4.05EE104 pKa = 4.03EE105 pKa = 3.86PTGIDD110 pKa = 2.72IGFGPGIVMPSVSNHH125 pKa = 5.61EE126 pKa = 4.11GGTYY130 pKa = 9.91VRR132 pKa = 11.84YY133 pKa = 10.17NGLGGIDD140 pKa = 4.39PNCKK144 pKa = 10.08DD145 pKa = 5.09LISKK149 pKa = 7.7MMRR152 pKa = 11.84SLIGQIGNKK161 pKa = 9.32YY162 pKa = 10.51GYY164 pKa = 10.63DD165 pKa = 3.29IDD167 pKa = 5.26LFDD170 pKa = 3.78YY171 pKa = 11.0QGDD174 pKa = 3.78FLEE177 pKa = 4.56VFLPHH182 pKa = 6.85KK183 pKa = 9.94PSKK186 pKa = 10.61EE187 pKa = 3.82DD188 pKa = 3.1VRR190 pKa = 11.84PDD192 pKa = 2.86IRR194 pKa = 11.84IGKK197 pKa = 9.19KK198 pKa = 9.77NEE200 pKa = 3.82EE201 pKa = 4.3GTSKK205 pKa = 10.38QVSKK209 pKa = 10.55PRR211 pKa = 11.84EE212 pKa = 3.73KK213 pKa = 10.86EE214 pKa = 3.94KK215 pKa = 10.77IVLKK219 pKa = 10.19TGDD222 pKa = 3.28EE223 pKa = 4.34CGRR226 pKa = 11.84FPMNKK231 pKa = 8.37EE232 pKa = 3.59AKK234 pKa = 9.41KK235 pKa = 10.55RR236 pKa = 11.84EE237 pKa = 4.09PEE239 pKa = 4.18GLWEE243 pKa = 4.12VMKK246 pKa = 10.93VLSVQFDD253 pKa = 3.36PWKK256 pKa = 10.41EE257 pKa = 3.96DD258 pKa = 3.66EE259 pKa = 5.12PPLSLTIRR267 pKa = 11.84DD268 pKa = 3.82LFISEE273 pKa = 4.35SEE275 pKa = 4.08FRR277 pKa = 11.84LHH279 pKa = 6.92CNHH282 pKa = 6.13SQTEE286 pKa = 4.08RR287 pKa = 11.84EE288 pKa = 3.94MALVGIKK295 pKa = 10.07LRR297 pKa = 11.84RR298 pKa = 11.84LYY300 pKa = 11.05NKK302 pKa = 10.09LYY304 pKa = 10.24QKK306 pKa = 10.81YY307 pKa = 9.63RR308 pKa = 11.84LL309 pKa = 3.67
Molecular weight: 35.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q88415|Q88415_SVCVA Glycoprotein OS=Spring viraemia of carp virus (strain ATCC VR-1390) OX=1559360 GN=G PE=4 SV=1
MM1 pKa = 6.82STLRR5 pKa = 11.84KK6 pKa = 10.05LFGTKK11 pKa = 9.43KK12 pKa = 10.68SKK14 pKa = 8.95GTPPTYY20 pKa = 10.74EE21 pKa = 4.01EE22 pKa = 4.31TLATAPVLMDD32 pKa = 2.99THH34 pKa = 6.8DD35 pKa = 3.75THH37 pKa = 7.47SHH39 pKa = 4.98SLQWMRR45 pKa = 11.84YY46 pKa = 7.96HH47 pKa = 7.71VEE49 pKa = 4.16LDD51 pKa = 3.69VKK53 pKa = 11.05LDD55 pKa = 3.82TPLKK59 pKa = 9.96TMSDD63 pKa = 3.5LLGLLKK69 pKa = 10.79NWDD72 pKa = 3.16VDD74 pKa = 4.11YY75 pKa = 11.22KK76 pKa = 10.99GSRR79 pKa = 11.84NKK81 pKa = 10.08RR82 pKa = 11.84RR83 pKa = 11.84FYY85 pKa = 10.36RR86 pKa = 11.84LIMFRR91 pKa = 11.84CALEE95 pKa = 4.25LKK97 pKa = 9.73HH98 pKa = 7.61VSGTYY103 pKa = 10.34SVDD106 pKa = 3.04GSALYY111 pKa = 10.53SNKK114 pKa = 9.44VQGSCYY120 pKa = 8.93VPHH123 pKa = 7.7RR124 pKa = 11.84FGQMPPFKK132 pKa = 10.6RR133 pKa = 11.84EE134 pKa = 3.71IEE136 pKa = 4.18VFRR139 pKa = 11.84YY140 pKa = 8.67PVHH143 pKa = 4.51QHH145 pKa = 5.98GYY147 pKa = 10.25NGMVDD152 pKa = 4.26LRR154 pKa = 11.84MSICDD159 pKa = 3.82LNGEE163 pKa = 4.34KK164 pKa = 9.87IGLNLLKK171 pKa = 9.71EE172 pKa = 4.53CQVAHH177 pKa = 6.87PNHH180 pKa = 5.61FQKK183 pKa = 11.0YY184 pKa = 8.03LEE186 pKa = 4.23EE187 pKa = 4.94VGLEE191 pKa = 4.13AACSATGEE199 pKa = 4.48WILDD203 pKa = 3.37WTFPMPVDD211 pKa = 3.55VVPRR215 pKa = 11.84VPSLFMGDD223 pKa = 3.19
MM1 pKa = 6.82STLRR5 pKa = 11.84KK6 pKa = 10.05LFGTKK11 pKa = 9.43KK12 pKa = 10.68SKK14 pKa = 8.95GTPPTYY20 pKa = 10.74EE21 pKa = 4.01EE22 pKa = 4.31TLATAPVLMDD32 pKa = 2.99THH34 pKa = 6.8DD35 pKa = 3.75THH37 pKa = 7.47SHH39 pKa = 4.98SLQWMRR45 pKa = 11.84YY46 pKa = 7.96HH47 pKa = 7.71VEE49 pKa = 4.16LDD51 pKa = 3.69VKK53 pKa = 11.05LDD55 pKa = 3.82TPLKK59 pKa = 9.96TMSDD63 pKa = 3.5LLGLLKK69 pKa = 10.79NWDD72 pKa = 3.16VDD74 pKa = 4.11YY75 pKa = 11.22KK76 pKa = 10.99GSRR79 pKa = 11.84NKK81 pKa = 10.08RR82 pKa = 11.84RR83 pKa = 11.84FYY85 pKa = 10.36RR86 pKa = 11.84LIMFRR91 pKa = 11.84CALEE95 pKa = 4.25LKK97 pKa = 9.73HH98 pKa = 7.61VSGTYY103 pKa = 10.34SVDD106 pKa = 3.04GSALYY111 pKa = 10.53SNKK114 pKa = 9.44VQGSCYY120 pKa = 8.93VPHH123 pKa = 7.7RR124 pKa = 11.84FGQMPPFKK132 pKa = 10.6RR133 pKa = 11.84EE134 pKa = 3.71IEE136 pKa = 4.18VFRR139 pKa = 11.84YY140 pKa = 8.67PVHH143 pKa = 4.51QHH145 pKa = 5.98GYY147 pKa = 10.25NGMVDD152 pKa = 4.26LRR154 pKa = 11.84MSICDD159 pKa = 3.82LNGEE163 pKa = 4.34KK164 pKa = 9.87IGLNLLKK171 pKa = 9.71EE172 pKa = 4.53CQVAHH177 pKa = 6.87PNHH180 pKa = 5.61FQKK183 pKa = 11.0YY184 pKa = 8.03LEE186 pKa = 4.23EE187 pKa = 4.94VGLEE191 pKa = 4.13AACSATGEE199 pKa = 4.48WILDD203 pKa = 3.37WTFPMPVDD211 pKa = 3.55VVPRR215 pKa = 11.84VPSLFMGDD223 pKa = 3.19
Molecular weight: 25.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3554 |
223 |
2095 |
710.8 |
80.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.249 ± 0.733 | 1.829 ± 0.267 |
5.684 ± 0.736 | 7.006 ± 0.891 |
3.545 ± 0.146 | 6.725 ± 0.4 |
2.842 ± 0.366 | 6.697 ± 0.758 |
6.331 ± 0.577 | 9.398 ± 0.799 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.814 ± 0.244 | 4.164 ± 0.38 |
4.502 ± 0.338 | 3.236 ± 0.347 |
5.346 ± 0.401 | 8.357 ± 0.668 |
5.74 ± 0.427 | 5.993 ± 0.619 |
1.998 ± 0.139 | 3.545 ± 0.281 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
