
Tupaia virus (isolate Tupaia/Thailand/-/1986) (TUPV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Tupavirus; Tupaia tupavirus; Tupaia virus
Average proteome isoelectric point is 7.14
Get precalculated fractions of proteins
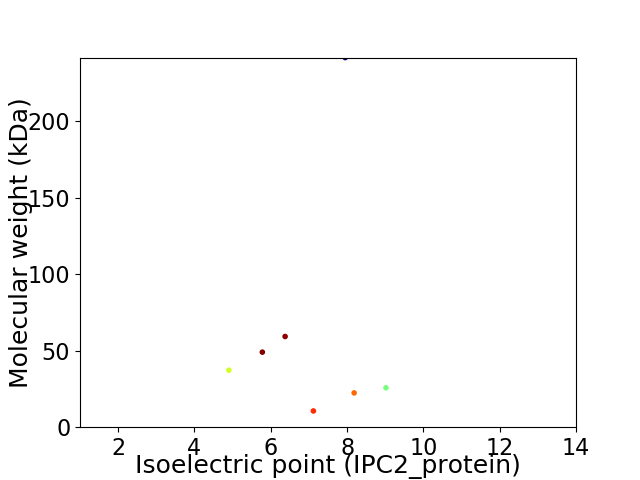
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q4VKV8|NCAP_TUPVT Nucleoprotein OS=Tupaia virus (isolate Tupaia/Thailand/-/1986) OX=1560034 GN=N PE=3 SV=1
MM1 pKa = 7.91TDD3 pKa = 3.16PKK5 pKa = 10.55KK6 pKa = 10.35ISRR9 pKa = 11.84VALKK13 pKa = 10.56GYY15 pKa = 10.64DD16 pKa = 3.37LTKK19 pKa = 10.46VSRR22 pKa = 11.84ALEE25 pKa = 4.13EE26 pKa = 5.54CEE28 pKa = 4.06DD29 pKa = 3.77QPGVRR34 pKa = 11.84VGGVQIVSGGEE45 pKa = 3.81AEE47 pKa = 4.69APAHH51 pKa = 6.93LEE53 pKa = 4.05DD54 pKa = 4.25NRR56 pKa = 11.84QGAIGDD62 pKa = 3.76NFDD65 pKa = 3.29VFSRR69 pKa = 11.84DD70 pKa = 3.26SKK72 pKa = 10.72NVQIVTEE79 pKa = 4.68HH80 pKa = 6.91EE81 pKa = 4.26DD82 pKa = 3.54SSDD85 pKa = 3.57EE86 pKa = 4.2EE87 pKa = 4.5YY88 pKa = 11.26EE89 pKa = 4.07EE90 pKa = 4.2ARR92 pKa = 11.84SYY94 pKa = 11.87GDD96 pKa = 4.87DD97 pKa = 4.41SGHH100 pKa = 5.43QPNRR104 pKa = 11.84AAVPFADD111 pKa = 5.43DD112 pKa = 3.96EE113 pKa = 4.67YY114 pKa = 11.62GDD116 pKa = 3.72FKK118 pKa = 10.78RR119 pKa = 11.84ARR121 pKa = 11.84EE122 pKa = 3.83RR123 pKa = 11.84LPQLFHH129 pKa = 6.81EE130 pKa = 5.31GWGFGSQQANEE141 pKa = 4.15EE142 pKa = 4.29SSEE145 pKa = 3.95GLQQQSGEE153 pKa = 4.31CGSGGGSSNAGGKK166 pKa = 10.01DD167 pKa = 3.35EE168 pKa = 6.87DD169 pKa = 4.63DD170 pKa = 4.02LQEE173 pKa = 4.18KK174 pKa = 8.85TGHH177 pKa = 5.66EE178 pKa = 4.22KK179 pKa = 10.33EE180 pKa = 4.39KK181 pKa = 11.16SVIVPPKK188 pKa = 9.66EE189 pKa = 4.02SGSVSGYY196 pKa = 10.28HH197 pKa = 6.53GSCDD201 pKa = 2.8IPFNMEE207 pKa = 3.92EE208 pKa = 4.35FLASPATTQRR218 pKa = 11.84RR219 pKa = 11.84QLLEE223 pKa = 4.25LCQLIADD230 pKa = 4.81RR231 pKa = 11.84SNEE234 pKa = 3.78EE235 pKa = 5.01LIFFPWGFNLVKK247 pKa = 10.44RR248 pKa = 11.84KK249 pKa = 6.48VHH251 pKa = 5.86RR252 pKa = 11.84VEE254 pKa = 3.96PQQPVAVSHH263 pKa = 6.62RR264 pKa = 11.84FTWEE268 pKa = 3.71DD269 pKa = 3.28FQLKK273 pKa = 9.91LKK275 pKa = 10.96AGFTLIHH282 pKa = 7.01KK283 pKa = 7.47KK284 pKa = 8.61TKK286 pKa = 10.68APVVLNSSSYY296 pKa = 10.63NLGSVPEE303 pKa = 4.38GGISLSPDD311 pKa = 3.22DD312 pKa = 4.6TEE314 pKa = 4.75LSVLIKK320 pKa = 10.16CLRR323 pKa = 11.84YY324 pKa = 9.51LGLYY328 pKa = 9.71KK329 pKa = 10.48FLATQIEE336 pKa = 4.55FF337 pKa = 3.68
MM1 pKa = 7.91TDD3 pKa = 3.16PKK5 pKa = 10.55KK6 pKa = 10.35ISRR9 pKa = 11.84VALKK13 pKa = 10.56GYY15 pKa = 10.64DD16 pKa = 3.37LTKK19 pKa = 10.46VSRR22 pKa = 11.84ALEE25 pKa = 4.13EE26 pKa = 5.54CEE28 pKa = 4.06DD29 pKa = 3.77QPGVRR34 pKa = 11.84VGGVQIVSGGEE45 pKa = 3.81AEE47 pKa = 4.69APAHH51 pKa = 6.93LEE53 pKa = 4.05DD54 pKa = 4.25NRR56 pKa = 11.84QGAIGDD62 pKa = 3.76NFDD65 pKa = 3.29VFSRR69 pKa = 11.84DD70 pKa = 3.26SKK72 pKa = 10.72NVQIVTEE79 pKa = 4.68HH80 pKa = 6.91EE81 pKa = 4.26DD82 pKa = 3.54SSDD85 pKa = 3.57EE86 pKa = 4.2EE87 pKa = 4.5YY88 pKa = 11.26EE89 pKa = 4.07EE90 pKa = 4.2ARR92 pKa = 11.84SYY94 pKa = 11.87GDD96 pKa = 4.87DD97 pKa = 4.41SGHH100 pKa = 5.43QPNRR104 pKa = 11.84AAVPFADD111 pKa = 5.43DD112 pKa = 3.96EE113 pKa = 4.67YY114 pKa = 11.62GDD116 pKa = 3.72FKK118 pKa = 10.78RR119 pKa = 11.84ARR121 pKa = 11.84EE122 pKa = 3.83RR123 pKa = 11.84LPQLFHH129 pKa = 6.81EE130 pKa = 5.31GWGFGSQQANEE141 pKa = 4.15EE142 pKa = 4.29SSEE145 pKa = 3.95GLQQQSGEE153 pKa = 4.31CGSGGGSSNAGGKK166 pKa = 10.01DD167 pKa = 3.35EE168 pKa = 6.87DD169 pKa = 4.63DD170 pKa = 4.02LQEE173 pKa = 4.18KK174 pKa = 8.85TGHH177 pKa = 5.66EE178 pKa = 4.22KK179 pKa = 10.33EE180 pKa = 4.39KK181 pKa = 11.16SVIVPPKK188 pKa = 9.66EE189 pKa = 4.02SGSVSGYY196 pKa = 10.28HH197 pKa = 6.53GSCDD201 pKa = 2.8IPFNMEE207 pKa = 3.92EE208 pKa = 4.35FLASPATTQRR218 pKa = 11.84RR219 pKa = 11.84QLLEE223 pKa = 4.25LCQLIADD230 pKa = 4.81RR231 pKa = 11.84SNEE234 pKa = 3.78EE235 pKa = 5.01LIFFPWGFNLVKK247 pKa = 10.44RR248 pKa = 11.84KK249 pKa = 6.48VHH251 pKa = 5.86RR252 pKa = 11.84VEE254 pKa = 3.96PQQPVAVSHH263 pKa = 6.62RR264 pKa = 11.84FTWEE268 pKa = 3.71DD269 pKa = 3.28FQLKK273 pKa = 9.91LKK275 pKa = 10.96AGFTLIHH282 pKa = 7.01KK283 pKa = 7.47KK284 pKa = 8.61TKK286 pKa = 10.68APVVLNSSSYY296 pKa = 10.63NLGSVPEE303 pKa = 4.38GGISLSPDD311 pKa = 3.22DD312 pKa = 4.6TEE314 pKa = 4.75LSVLIKK320 pKa = 10.16CLRR323 pKa = 11.84YY324 pKa = 9.51LGLYY328 pKa = 9.71KK329 pKa = 10.48FLATQIEE336 pKa = 4.55FF337 pKa = 3.68
Molecular weight: 37.29 kDa
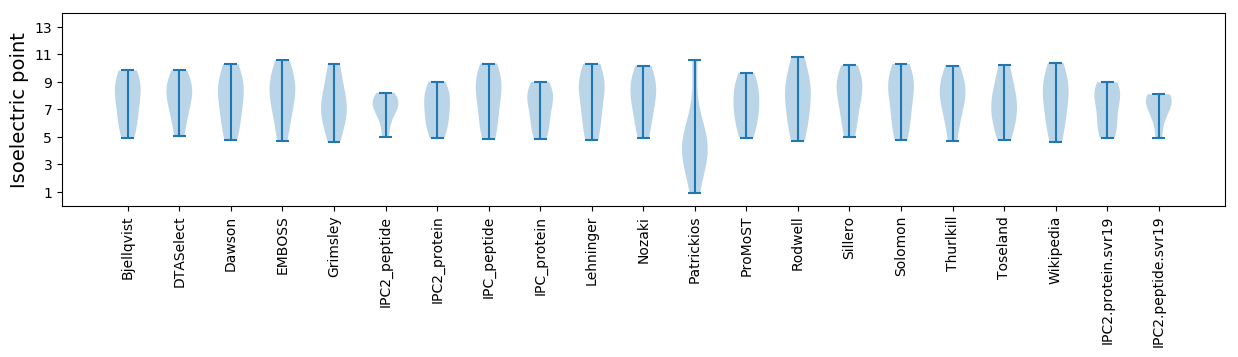
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q4VKV7|PHOSP_TUPVT Phosphoprotein OS=Tupaia virus (isolate Tupaia/Thailand/-/1986) OX=1560034 GN=P PE=3 SV=1
MM1 pKa = 7.24TSQRR5 pKa = 11.84FQEE8 pKa = 3.98LLKK11 pKa = 10.83NVKK14 pKa = 9.15TNLGSVWVEE23 pKa = 3.55FRR25 pKa = 11.84SSVEE29 pKa = 3.63EE30 pKa = 4.1RR31 pKa = 11.84LKK33 pKa = 10.97HH34 pKa = 4.12PHH36 pKa = 4.87IWKK39 pKa = 10.11IIDD42 pKa = 3.49RR43 pKa = 11.84EE44 pKa = 4.11QLATILMFFLGIPKK58 pKa = 9.3MFRR61 pKa = 11.84LSQNMKK67 pKa = 8.95IRR69 pKa = 11.84QMRR72 pKa = 11.84NMRR75 pKa = 11.84KK76 pKa = 8.7PGVMEE81 pKa = 3.8MTPDD85 pKa = 3.56TNQTEE90 pKa = 4.61QPCPSQMTNTEE101 pKa = 4.02TSKK104 pKa = 10.84EE105 pKa = 4.13PEE107 pKa = 4.25SVSLNSSTKK116 pKa = 10.98DD117 pKa = 3.21GDD119 pKa = 3.98LDD121 pKa = 4.24PNKK124 pKa = 10.17QMKK127 pKa = 9.59NLRR130 pKa = 11.84RR131 pKa = 11.84DD132 pKa = 3.23SSSSQEE138 pKa = 4.22SADD141 pKa = 3.69LEE143 pKa = 4.28EE144 pKa = 4.42DD145 pKa = 3.01HH146 pKa = 7.2RR147 pKa = 11.84MLGGRR152 pKa = 11.84MRR154 pKa = 11.84MIFKK158 pKa = 10.22RR159 pKa = 11.84RR160 pKa = 11.84QGMRR164 pKa = 11.84KK165 pKa = 9.13KK166 pKa = 10.2SQSLSHH172 pKa = 6.22LRR174 pKa = 11.84SQAVYY179 pKa = 10.8QDD181 pKa = 3.5TTEE184 pKa = 4.05AAISPSTWKK193 pKa = 10.87SFWPPQLRR201 pKa = 11.84LRR203 pKa = 11.84DD204 pKa = 4.01DD205 pKa = 4.25NSWSSVNLSQTDD217 pKa = 3.53PTKK220 pKa = 11.15NN221 pKa = 3.09
MM1 pKa = 7.24TSQRR5 pKa = 11.84FQEE8 pKa = 3.98LLKK11 pKa = 10.83NVKK14 pKa = 9.15TNLGSVWVEE23 pKa = 3.55FRR25 pKa = 11.84SSVEE29 pKa = 3.63EE30 pKa = 4.1RR31 pKa = 11.84LKK33 pKa = 10.97HH34 pKa = 4.12PHH36 pKa = 4.87IWKK39 pKa = 10.11IIDD42 pKa = 3.49RR43 pKa = 11.84EE44 pKa = 4.11QLATILMFFLGIPKK58 pKa = 9.3MFRR61 pKa = 11.84LSQNMKK67 pKa = 8.95IRR69 pKa = 11.84QMRR72 pKa = 11.84NMRR75 pKa = 11.84KK76 pKa = 8.7PGVMEE81 pKa = 3.8MTPDD85 pKa = 3.56TNQTEE90 pKa = 4.61QPCPSQMTNTEE101 pKa = 4.02TSKK104 pKa = 10.84EE105 pKa = 4.13PEE107 pKa = 4.25SVSLNSSTKK116 pKa = 10.98DD117 pKa = 3.21GDD119 pKa = 3.98LDD121 pKa = 4.24PNKK124 pKa = 10.17QMKK127 pKa = 9.59NLRR130 pKa = 11.84RR131 pKa = 11.84DD132 pKa = 3.23SSSSQEE138 pKa = 4.22SADD141 pKa = 3.69LEE143 pKa = 4.28EE144 pKa = 4.42DD145 pKa = 3.01HH146 pKa = 7.2RR147 pKa = 11.84MLGGRR152 pKa = 11.84MRR154 pKa = 11.84MIFKK158 pKa = 10.22RR159 pKa = 11.84RR160 pKa = 11.84QGMRR164 pKa = 11.84KK165 pKa = 9.13KK166 pKa = 10.2SQSLSHH172 pKa = 6.22LRR174 pKa = 11.84SQAVYY179 pKa = 10.8QDD181 pKa = 3.5TTEE184 pKa = 4.05AAISPSTWKK193 pKa = 10.87SFWPPQLRR201 pKa = 11.84LRR203 pKa = 11.84DD204 pKa = 4.01DD205 pKa = 4.25NSWSSVNLSQTDD217 pKa = 3.53PTKK220 pKa = 11.15NN221 pKa = 3.09
Molecular weight: 25.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3919 |
93 |
2107 |
559.9 |
63.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.44 ± 0.494 | 1.684 ± 0.265 |
5.307 ± 0.782 | 6.098 ± 0.709 |
5.078 ± 0.663 | 6.047 ± 0.592 |
2.194 ± 0.197 | 6.073 ± 0.617 |
6.022 ± 0.398 | 10.794 ± 1.157 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.067 ± 0.395 | 4.389 ± 0.21 |
4.746 ± 0.125 | 3.521 ± 0.57 |
5.792 ± 0.742 | 8.829 ± 0.373 |
5.639 ± 0.753 | 6.073 ± 0.545 |
1.99 ± 0.219 | 3.215 ± 0.316 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
