
Nitrosospira sp. Nsp14
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Nitrosomonadales; Nitrosomonadaceae; Nitrosospira; unclassified Nitrosospira
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
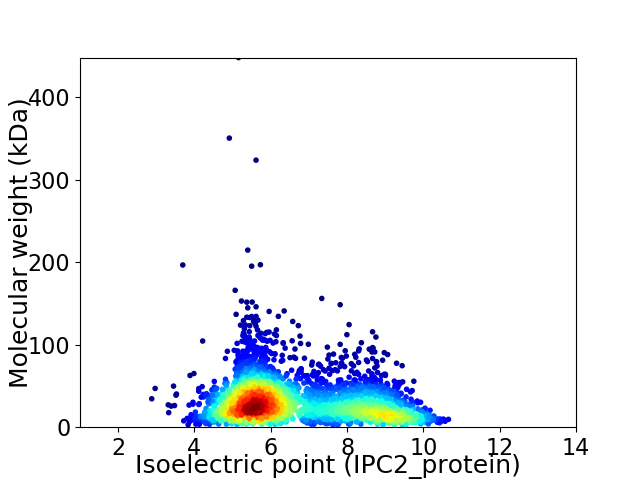
Virtual 2D-PAGE plot for 3081 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I3AYG6|A0A1I3AYG6_9PROT N-acyl homoserine lactone hydrolase OS=Nitrosospira sp. Nsp14 OX=1855333 GN=SAMN05216299_12130 PE=4 SV=1
MM1 pKa = 7.33SSVYY5 pKa = 10.78SSGTYY10 pKa = 6.59TTVGPAPTGNSTINALIVGTKK31 pKa = 9.0WGASGLGGSANVTFSFPVSFTVGSSGAHH59 pKa = 4.87SSGAFGYY66 pKa = 8.9TGEE69 pKa = 4.64PQDD72 pKa = 4.11SGQALSAMQMAAARR86 pKa = 11.84EE87 pKa = 4.17TLHH90 pKa = 5.73KK91 pKa = 8.85WSRR94 pKa = 11.84VATLNCNEE102 pKa = 4.12VADD105 pKa = 4.76TIDD108 pKa = 3.86TKK110 pKa = 10.82WEE112 pKa = 4.05SGDD115 pKa = 3.52NSSVGDD121 pKa = 3.54IRR123 pKa = 11.84LAMSNLPSAAWAYY136 pKa = 11.15YY137 pKa = 9.15PGQDD141 pKa = 3.26NAPYY145 pKa = 10.74GGDD148 pKa = 2.89VWLNKK153 pKa = 9.75SDD155 pKa = 3.56YY156 pKa = 9.35HH157 pKa = 7.86DD158 pKa = 4.17PVVGYY163 pKa = 8.49YY164 pKa = 10.36AYY166 pKa = 9.65YY167 pKa = 10.25TFLHH171 pKa = 6.78EE172 pKa = 4.76IGHH175 pKa = 5.3TLGLEE180 pKa = 4.38HH181 pKa = 6.79PHH183 pKa = 6.68EE184 pKa = 5.22SNDD187 pKa = 3.41NDD189 pKa = 3.9VMSLSVDD196 pKa = 3.17ALKK199 pKa = 11.01YY200 pKa = 10.78SIMSYY205 pKa = 10.33RR206 pKa = 11.84DD207 pKa = 3.39YY208 pKa = 11.57VGDD211 pKa = 5.14DD212 pKa = 2.96IDD214 pKa = 4.63GVGEE218 pKa = 4.21NFFPTTPMLYY228 pKa = 10.3DD229 pKa = 3.56VLALQALYY237 pKa = 10.1GANWNYY243 pKa = 10.71RR244 pKa = 11.84SGNDD248 pKa = 3.37TYY250 pKa = 11.74SWAPGAKK257 pKa = 9.39VYY259 pKa = 8.0EE260 pKa = 4.59TIWDD264 pKa = 3.63GGGIDD269 pKa = 4.0TLSAANQTRR278 pKa = 11.84DD279 pKa = 3.43VEE281 pKa = 4.28LHH283 pKa = 5.34LTSGVFSKK291 pKa = 10.42IGSPVWDD298 pKa = 3.38EE299 pKa = 4.26HH300 pKa = 7.34AWVRR304 pKa = 11.84DD305 pKa = 3.47NLVIAFNATIEE316 pKa = 4.16NATGSSYY323 pKa = 11.54DD324 pKa = 4.01DD325 pKa = 3.96YY326 pKa = 11.67IFGNTAGNVLVGGPGNDD343 pKa = 3.17TMRR346 pKa = 11.84GSEE349 pKa = 3.79WADD352 pKa = 2.8RR353 pKa = 11.84DD354 pKa = 3.98VFDD357 pKa = 5.14NDD359 pKa = 3.19TMYY362 pKa = 11.05GGSGGDD368 pKa = 3.97LMAGHH373 pKa = 6.9NGNDD377 pKa = 3.06VMYY380 pKa = 10.12GGEE383 pKa = 4.21GNDD386 pKa = 3.71FVTGDD391 pKa = 3.82SGNDD395 pKa = 3.02ILSGGNGNDD404 pKa = 3.51ILNGGPGDD412 pKa = 4.1DD413 pKa = 4.64LLIGGLGNDD422 pKa = 3.96TLNGGGGFDD431 pKa = 3.54EE432 pKa = 4.51ATYY435 pKa = 10.8AAATAPVNVDD445 pKa = 3.72LSLSGGRR452 pKa = 11.84ATGAQGIDD460 pKa = 3.2KK461 pKa = 10.61LRR463 pKa = 11.84GIEE466 pKa = 4.22KK467 pKa = 8.7VTGSAFADD475 pKa = 3.96TLTGDD480 pKa = 3.36ANANVLSGRR489 pKa = 11.84LGNDD493 pKa = 3.29TLQGGAGNDD502 pKa = 3.55TLAGGGGDD510 pKa = 5.48DD511 pKa = 4.31LFRR514 pKa = 11.84WTATAFNSGDD524 pKa = 3.16IRR526 pKa = 11.84FGGVDD531 pKa = 3.12RR532 pKa = 11.84VSDD535 pKa = 4.33FSRR538 pKa = 11.84GDD540 pKa = 3.47ALDD543 pKa = 4.48FSAALEE549 pKa = 4.07SLLMVNGTRR558 pKa = 11.84LGAATSDD565 pKa = 3.22IALGNALSISTNVCLQNGRR584 pKa = 11.84DD585 pKa = 3.87LYY587 pKa = 10.84IDD589 pKa = 4.6LDD591 pKa = 3.97SSHH594 pKa = 7.21SITANDD600 pKa = 3.44YY601 pKa = 9.95HH602 pKa = 8.82VEE604 pKa = 4.01LTGLGGVLKK613 pKa = 10.58YY614 pKa = 10.59DD615 pKa = 3.52AAADD619 pKa = 3.94LFHH622 pKa = 6.76VV623 pKa = 4.49
MM1 pKa = 7.33SSVYY5 pKa = 10.78SSGTYY10 pKa = 6.59TTVGPAPTGNSTINALIVGTKK31 pKa = 9.0WGASGLGGSANVTFSFPVSFTVGSSGAHH59 pKa = 4.87SSGAFGYY66 pKa = 8.9TGEE69 pKa = 4.64PQDD72 pKa = 4.11SGQALSAMQMAAARR86 pKa = 11.84EE87 pKa = 4.17TLHH90 pKa = 5.73KK91 pKa = 8.85WSRR94 pKa = 11.84VATLNCNEE102 pKa = 4.12VADD105 pKa = 4.76TIDD108 pKa = 3.86TKK110 pKa = 10.82WEE112 pKa = 4.05SGDD115 pKa = 3.52NSSVGDD121 pKa = 3.54IRR123 pKa = 11.84LAMSNLPSAAWAYY136 pKa = 11.15YY137 pKa = 9.15PGQDD141 pKa = 3.26NAPYY145 pKa = 10.74GGDD148 pKa = 2.89VWLNKK153 pKa = 9.75SDD155 pKa = 3.56YY156 pKa = 9.35HH157 pKa = 7.86DD158 pKa = 4.17PVVGYY163 pKa = 8.49YY164 pKa = 10.36AYY166 pKa = 9.65YY167 pKa = 10.25TFLHH171 pKa = 6.78EE172 pKa = 4.76IGHH175 pKa = 5.3TLGLEE180 pKa = 4.38HH181 pKa = 6.79PHH183 pKa = 6.68EE184 pKa = 5.22SNDD187 pKa = 3.41NDD189 pKa = 3.9VMSLSVDD196 pKa = 3.17ALKK199 pKa = 11.01YY200 pKa = 10.78SIMSYY205 pKa = 10.33RR206 pKa = 11.84DD207 pKa = 3.39YY208 pKa = 11.57VGDD211 pKa = 5.14DD212 pKa = 2.96IDD214 pKa = 4.63GVGEE218 pKa = 4.21NFFPTTPMLYY228 pKa = 10.3DD229 pKa = 3.56VLALQALYY237 pKa = 10.1GANWNYY243 pKa = 10.71RR244 pKa = 11.84SGNDD248 pKa = 3.37TYY250 pKa = 11.74SWAPGAKK257 pKa = 9.39VYY259 pKa = 8.0EE260 pKa = 4.59TIWDD264 pKa = 3.63GGGIDD269 pKa = 4.0TLSAANQTRR278 pKa = 11.84DD279 pKa = 3.43VEE281 pKa = 4.28LHH283 pKa = 5.34LTSGVFSKK291 pKa = 10.42IGSPVWDD298 pKa = 3.38EE299 pKa = 4.26HH300 pKa = 7.34AWVRR304 pKa = 11.84DD305 pKa = 3.47NLVIAFNATIEE316 pKa = 4.16NATGSSYY323 pKa = 11.54DD324 pKa = 4.01DD325 pKa = 3.96YY326 pKa = 11.67IFGNTAGNVLVGGPGNDD343 pKa = 3.17TMRR346 pKa = 11.84GSEE349 pKa = 3.79WADD352 pKa = 2.8RR353 pKa = 11.84DD354 pKa = 3.98VFDD357 pKa = 5.14NDD359 pKa = 3.19TMYY362 pKa = 11.05GGSGGDD368 pKa = 3.97LMAGHH373 pKa = 6.9NGNDD377 pKa = 3.06VMYY380 pKa = 10.12GGEE383 pKa = 4.21GNDD386 pKa = 3.71FVTGDD391 pKa = 3.82SGNDD395 pKa = 3.02ILSGGNGNDD404 pKa = 3.51ILNGGPGDD412 pKa = 4.1DD413 pKa = 4.64LLIGGLGNDD422 pKa = 3.96TLNGGGGFDD431 pKa = 3.54EE432 pKa = 4.51ATYY435 pKa = 10.8AAATAPVNVDD445 pKa = 3.72LSLSGGRR452 pKa = 11.84ATGAQGIDD460 pKa = 3.2KK461 pKa = 10.61LRR463 pKa = 11.84GIEE466 pKa = 4.22KK467 pKa = 8.7VTGSAFADD475 pKa = 3.96TLTGDD480 pKa = 3.36ANANVLSGRR489 pKa = 11.84LGNDD493 pKa = 3.29TLQGGAGNDD502 pKa = 3.55TLAGGGGDD510 pKa = 5.48DD511 pKa = 4.31LFRR514 pKa = 11.84WTATAFNSGDD524 pKa = 3.16IRR526 pKa = 11.84FGGVDD531 pKa = 3.12RR532 pKa = 11.84VSDD535 pKa = 4.33FSRR538 pKa = 11.84GDD540 pKa = 3.47ALDD543 pKa = 4.48FSAALEE549 pKa = 4.07SLLMVNGTRR558 pKa = 11.84LGAATSDD565 pKa = 3.22IALGNALSISTNVCLQNGRR584 pKa = 11.84DD585 pKa = 3.87LYY587 pKa = 10.84IDD589 pKa = 4.6LDD591 pKa = 3.97SSHH594 pKa = 7.21SITANDD600 pKa = 3.44YY601 pKa = 9.95HH602 pKa = 8.82VEE604 pKa = 4.01LTGLGGVLKK613 pKa = 10.58YY614 pKa = 10.59DD615 pKa = 3.52AAADD619 pKa = 3.94LFHH622 pKa = 6.76VV623 pKa = 4.49
Molecular weight: 65.06 kDa
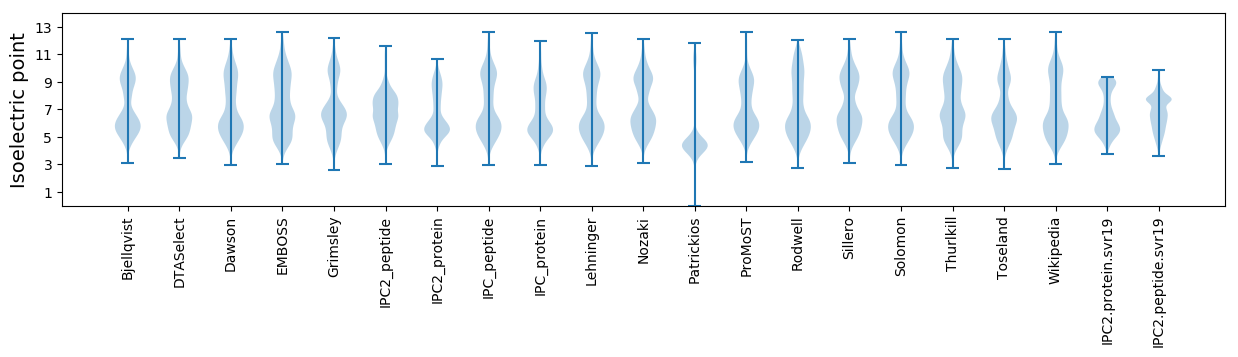
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I2ZTJ4|A0A1I2ZTJ4_9PROT ATP-dependent Clp protease ATP-binding subunit ClpA OS=Nitrosospira sp. Nsp14 OX=1855333 GN=SAMN05216299_11138 PE=3 SV=1
MM1 pKa = 7.9SPRR4 pKa = 11.84LTNNYY9 pKa = 7.63TLHH12 pKa = 6.21KK13 pKa = 7.15TTHH16 pKa = 4.32VWKK19 pKa = 10.53KK20 pKa = 9.05LQCQYY25 pKa = 10.91GCRR28 pKa = 11.84DD29 pKa = 3.52LRR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 9.78INARR37 pKa = 11.84GAIGVMAMVRR47 pKa = 11.84VEE49 pKa = 3.78RR50 pKa = 11.84STRR53 pKa = 11.84RR54 pKa = 11.84QEE56 pKa = 3.93ILDD59 pKa = 3.37YY60 pKa = 10.0WVRR63 pKa = 11.84ADD65 pKa = 3.5SVIIEE70 pKa = 3.96FRR72 pKa = 11.84PPGRR76 pKa = 11.84YY77 pKa = 9.2CSVFAAILILAA88 pKa = 4.82
MM1 pKa = 7.9SPRR4 pKa = 11.84LTNNYY9 pKa = 7.63TLHH12 pKa = 6.21KK13 pKa = 7.15TTHH16 pKa = 4.32VWKK19 pKa = 10.53KK20 pKa = 9.05LQCQYY25 pKa = 10.91GCRR28 pKa = 11.84DD29 pKa = 3.52LRR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 9.78INARR37 pKa = 11.84GAIGVMAMVRR47 pKa = 11.84VEE49 pKa = 3.78RR50 pKa = 11.84STRR53 pKa = 11.84RR54 pKa = 11.84QEE56 pKa = 3.93ILDD59 pKa = 3.37YY60 pKa = 10.0WVRR63 pKa = 11.84ADD65 pKa = 3.5SVIIEE70 pKa = 3.96FRR72 pKa = 11.84PPGRR76 pKa = 11.84YY77 pKa = 9.2CSVFAAILILAA88 pKa = 4.82
Molecular weight: 10.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
940644 |
25 |
4044 |
305.3 |
33.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.239 ± 0.054 | 0.967 ± 0.015 |
5.297 ± 0.036 | 5.977 ± 0.045 |
3.835 ± 0.035 | 8.007 ± 0.054 |
2.365 ± 0.021 | 5.583 ± 0.035 |
4.051 ± 0.04 | 10.42 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.468 ± 0.025 | 3.45 ± 0.034 |
4.881 ± 0.035 | 3.683 ± 0.027 |
6.518 ± 0.043 | 5.997 ± 0.033 |
5.149 ± 0.035 | 7.07 ± 0.038 |
1.323 ± 0.021 | 2.721 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
