
LeviOr01 phage
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Allassoviricetes; Levivirales; Leviviridae; unclassified Leviviridae
Average proteome isoelectric point is 8.14
Get precalculated fractions of proteins
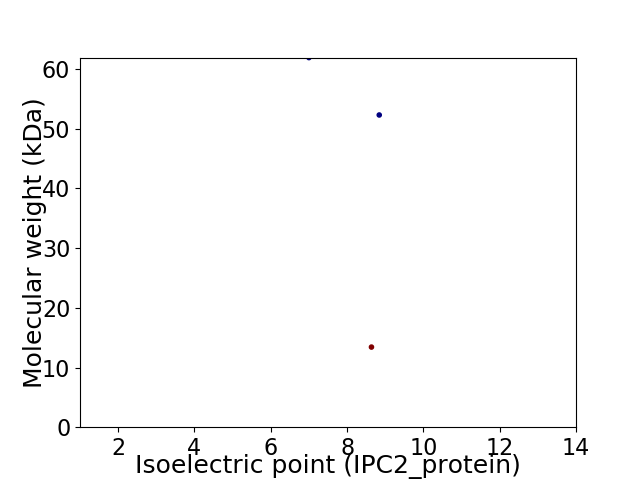
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1U6U245|A0A1U6U245_9VIRU Putative maturation protein OS=LeviOr01 phage OX=1927956 GN=mat PE=4 SV=1
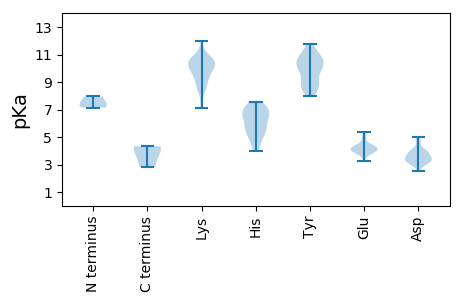
MM1 pKa = 7.13HH2 pKa = 7.56TLTAQVTSALLQDD15 pKa = 4.94LGLEE19 pKa = 4.19SLPKK23 pKa = 10.17EE24 pKa = 4.29FPDD27 pKa = 4.37RR28 pKa = 11.84DD29 pKa = 3.77SEE31 pKa = 4.61FPARR35 pKa = 11.84YY36 pKa = 8.49LAYY39 pKa = 10.34NFVRR43 pKa = 11.84KK44 pKa = 10.24LEE46 pKa = 4.24PFSRR50 pKa = 11.84KK51 pKa = 7.42TDD53 pKa = 3.26VPVAVLRR60 pKa = 11.84NSLTTFMEE68 pKa = 3.94AEE70 pKa = 4.05YY71 pKa = 10.21RR72 pKa = 11.84CRR74 pKa = 11.84VVNHH78 pKa = 6.67HH79 pKa = 5.58GRR81 pKa = 11.84MYY83 pKa = 10.45SALRR87 pKa = 11.84EE88 pKa = 3.85EE89 pKa = 4.33DD90 pKa = 2.81RR91 pKa = 11.84SYY93 pKa = 10.13FTEE96 pKa = 4.13AFRR99 pKa = 11.84LAKK102 pKa = 9.92IWINQTMSGYY112 pKa = 8.71WPRR115 pKa = 11.84WEE117 pKa = 3.92NARR120 pKa = 11.84YY121 pKa = 7.96TGGASRR127 pKa = 11.84NCSRR131 pKa = 11.84DD132 pKa = 3.01RR133 pKa = 11.84SLPALKK139 pKa = 9.86WSGYY143 pKa = 8.56AEE145 pKa = 4.08RR146 pKa = 11.84SNLSTTRR153 pKa = 11.84PALAVVNDD161 pKa = 3.89YY162 pKa = 10.78IKK164 pKa = 10.93PEE166 pKa = 4.17FSPSDD171 pKa = 3.08WRR173 pKa = 11.84EE174 pKa = 3.4RR175 pKa = 11.84DD176 pKa = 3.02VDD178 pKa = 3.62IVDD181 pKa = 3.94DD182 pKa = 3.97SRR184 pKa = 11.84FDD186 pKa = 3.98FVAKK190 pKa = 8.53TAKK193 pKa = 10.02AVRR196 pKa = 11.84FMAMEE201 pKa = 4.38PEE203 pKa = 4.23YY204 pKa = 11.8NMLAQKK210 pKa = 10.65CVGDD214 pKa = 4.75CIRR217 pKa = 11.84AALNAQGINLDD228 pKa = 3.73DD229 pKa = 3.56QRR231 pKa = 11.84PNQEE235 pKa = 3.63LAYY238 pKa = 9.89LGSIFRR244 pKa = 11.84TRR246 pKa = 11.84ATLDD250 pKa = 3.03QSSASDD256 pKa = 4.38CIALFLLRR264 pKa = 11.84LLPEE268 pKa = 4.37RR269 pKa = 11.84VRR271 pKa = 11.84DD272 pKa = 3.48WVLACRR278 pKa = 11.84TPATSVAGSRR288 pKa = 11.84LVLEE292 pKa = 5.25KK293 pKa = 10.72VATMGNGFIFEE304 pKa = 4.28LQSLIFAAFAHH315 pKa = 6.8ACTQLSGGRR324 pKa = 11.84EE325 pKa = 4.0CDD327 pKa = 2.97IAVYY331 pKa = 10.55GDD333 pKa = 4.73DD334 pKa = 4.84IIVSTPVARR343 pKa = 11.84PLMDD347 pKa = 3.35TLEE350 pKa = 4.32YY351 pKa = 10.85YY352 pKa = 11.01GLIPNMEE359 pKa = 4.02KK360 pKa = 10.51SYY362 pKa = 10.46WDD364 pKa = 3.13EE365 pKa = 4.4DD366 pKa = 3.83EE367 pKa = 5.05PFRR370 pKa = 11.84EE371 pKa = 4.44SCGKK375 pKa = 9.49HH376 pKa = 3.95WFAGRR381 pKa = 11.84DD382 pKa = 3.35VTPFYY387 pKa = 11.15VKK389 pKa = 10.55EE390 pKa = 3.87PLGPLRR396 pKa = 11.84TLFRR400 pKa = 11.84AYY402 pKa = 10.68NGLKK406 pKa = 8.99EE407 pKa = 3.85WTMRR411 pKa = 11.84TGIPLNRR418 pKa = 11.84TLATILAAIPKK429 pKa = 8.38KK430 pKa = 10.58DD431 pKa = 3.42RR432 pKa = 11.84VIVPPSFSIDD442 pKa = 3.74CGLHH446 pKa = 5.41TPVSGCTFPKK456 pKa = 10.0RR457 pKa = 11.84VIRR460 pKa = 11.84HH461 pKa = 4.55GDD463 pKa = 2.53IRR465 pKa = 11.84YY466 pKa = 9.76SFKK469 pKa = 11.08CLVDD473 pKa = 3.31TSEE476 pKa = 5.04DD477 pKa = 3.27VTARR481 pKa = 11.84LDD483 pKa = 3.83DD484 pKa = 4.2EE485 pKa = 4.89VKK487 pKa = 10.28LRR489 pKa = 11.84YY490 pKa = 8.71WLFEE494 pKa = 4.24PPAEE498 pKa = 3.95LLPRR502 pKa = 11.84HH503 pKa = 6.71LYY505 pKa = 8.88PASPLAGYY513 pKa = 8.97PRR515 pKa = 11.84EE516 pKa = 3.95EE517 pKa = 3.75KK518 pKa = 10.38RR519 pKa = 11.84YY520 pKa = 9.29VRR522 pKa = 11.84GLSGEE527 pKa = 4.11SRR529 pKa = 11.84PEE531 pKa = 3.23VWARR535 pKa = 11.84RR536 pKa = 11.84EE537 pKa = 4.2AGPGDD542 pKa = 3.74APP544 pKa = 4.18
MM1 pKa = 7.13HH2 pKa = 7.56TLTAQVTSALLQDD15 pKa = 4.94LGLEE19 pKa = 4.19SLPKK23 pKa = 10.17EE24 pKa = 4.29FPDD27 pKa = 4.37RR28 pKa = 11.84DD29 pKa = 3.77SEE31 pKa = 4.61FPARR35 pKa = 11.84YY36 pKa = 8.49LAYY39 pKa = 10.34NFVRR43 pKa = 11.84KK44 pKa = 10.24LEE46 pKa = 4.24PFSRR50 pKa = 11.84KK51 pKa = 7.42TDD53 pKa = 3.26VPVAVLRR60 pKa = 11.84NSLTTFMEE68 pKa = 3.94AEE70 pKa = 4.05YY71 pKa = 10.21RR72 pKa = 11.84CRR74 pKa = 11.84VVNHH78 pKa = 6.67HH79 pKa = 5.58GRR81 pKa = 11.84MYY83 pKa = 10.45SALRR87 pKa = 11.84EE88 pKa = 3.85EE89 pKa = 4.33DD90 pKa = 2.81RR91 pKa = 11.84SYY93 pKa = 10.13FTEE96 pKa = 4.13AFRR99 pKa = 11.84LAKK102 pKa = 9.92IWINQTMSGYY112 pKa = 8.71WPRR115 pKa = 11.84WEE117 pKa = 3.92NARR120 pKa = 11.84YY121 pKa = 7.96TGGASRR127 pKa = 11.84NCSRR131 pKa = 11.84DD132 pKa = 3.01RR133 pKa = 11.84SLPALKK139 pKa = 9.86WSGYY143 pKa = 8.56AEE145 pKa = 4.08RR146 pKa = 11.84SNLSTTRR153 pKa = 11.84PALAVVNDD161 pKa = 3.89YY162 pKa = 10.78IKK164 pKa = 10.93PEE166 pKa = 4.17FSPSDD171 pKa = 3.08WRR173 pKa = 11.84EE174 pKa = 3.4RR175 pKa = 11.84DD176 pKa = 3.02VDD178 pKa = 3.62IVDD181 pKa = 3.94DD182 pKa = 3.97SRR184 pKa = 11.84FDD186 pKa = 3.98FVAKK190 pKa = 8.53TAKK193 pKa = 10.02AVRR196 pKa = 11.84FMAMEE201 pKa = 4.38PEE203 pKa = 4.23YY204 pKa = 11.8NMLAQKK210 pKa = 10.65CVGDD214 pKa = 4.75CIRR217 pKa = 11.84AALNAQGINLDD228 pKa = 3.73DD229 pKa = 3.56QRR231 pKa = 11.84PNQEE235 pKa = 3.63LAYY238 pKa = 9.89LGSIFRR244 pKa = 11.84TRR246 pKa = 11.84ATLDD250 pKa = 3.03QSSASDD256 pKa = 4.38CIALFLLRR264 pKa = 11.84LLPEE268 pKa = 4.37RR269 pKa = 11.84VRR271 pKa = 11.84DD272 pKa = 3.48WVLACRR278 pKa = 11.84TPATSVAGSRR288 pKa = 11.84LVLEE292 pKa = 5.25KK293 pKa = 10.72VATMGNGFIFEE304 pKa = 4.28LQSLIFAAFAHH315 pKa = 6.8ACTQLSGGRR324 pKa = 11.84EE325 pKa = 4.0CDD327 pKa = 2.97IAVYY331 pKa = 10.55GDD333 pKa = 4.73DD334 pKa = 4.84IIVSTPVARR343 pKa = 11.84PLMDD347 pKa = 3.35TLEE350 pKa = 4.32YY351 pKa = 10.85YY352 pKa = 11.01GLIPNMEE359 pKa = 4.02KK360 pKa = 10.51SYY362 pKa = 10.46WDD364 pKa = 3.13EE365 pKa = 4.4DD366 pKa = 3.83EE367 pKa = 5.05PFRR370 pKa = 11.84EE371 pKa = 4.44SCGKK375 pKa = 9.49HH376 pKa = 3.95WFAGRR381 pKa = 11.84DD382 pKa = 3.35VTPFYY387 pKa = 11.15VKK389 pKa = 10.55EE390 pKa = 3.87PLGPLRR396 pKa = 11.84TLFRR400 pKa = 11.84AYY402 pKa = 10.68NGLKK406 pKa = 8.99EE407 pKa = 3.85WTMRR411 pKa = 11.84TGIPLNRR418 pKa = 11.84TLATILAAIPKK429 pKa = 8.38KK430 pKa = 10.58DD431 pKa = 3.42RR432 pKa = 11.84VIVPPSFSIDD442 pKa = 3.74CGLHH446 pKa = 5.41TPVSGCTFPKK456 pKa = 10.0RR457 pKa = 11.84VIRR460 pKa = 11.84HH461 pKa = 4.55GDD463 pKa = 2.53IRR465 pKa = 11.84YY466 pKa = 9.76SFKK469 pKa = 11.08CLVDD473 pKa = 3.31TSEE476 pKa = 5.04DD477 pKa = 3.27VTARR481 pKa = 11.84LDD483 pKa = 3.83DD484 pKa = 4.2EE485 pKa = 4.89VKK487 pKa = 10.28LRR489 pKa = 11.84YY490 pKa = 8.71WLFEE494 pKa = 4.24PPAEE498 pKa = 3.95LLPRR502 pKa = 11.84HH503 pKa = 6.71LYY505 pKa = 8.88PASPLAGYY513 pKa = 8.97PRR515 pKa = 11.84EE516 pKa = 3.95EE517 pKa = 3.75KK518 pKa = 10.38RR519 pKa = 11.84YY520 pKa = 9.29VRR522 pKa = 11.84GLSGEE527 pKa = 4.11SRR529 pKa = 11.84PEE531 pKa = 3.23VWARR535 pKa = 11.84RR536 pKa = 11.84EE537 pKa = 4.2AGPGDD542 pKa = 3.74APP544 pKa = 4.18
Molecular weight: 61.91 kDa
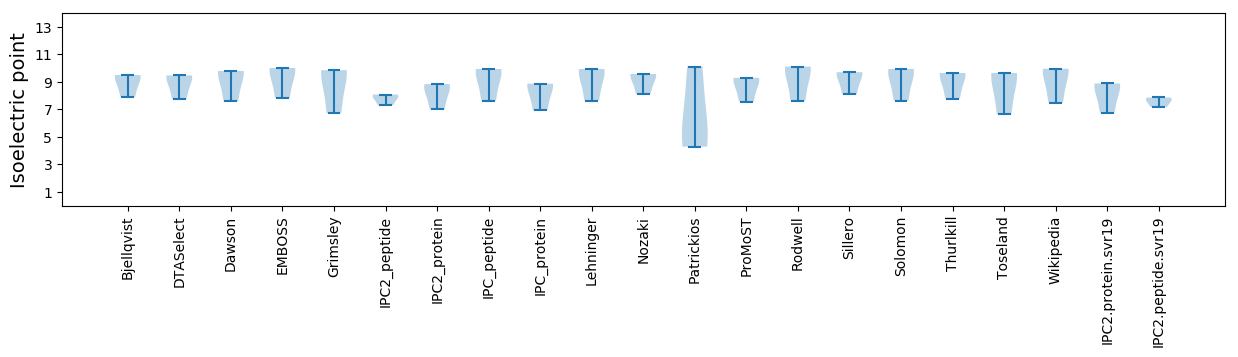
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1U6U260|A0A1U6U260_9VIRU Putative coat protein OS=LeviOr01 phage OX=1927956 GN=cp PE=4 SV=1
MM1 pKa = 7.95RR2 pKa = 11.84SEE4 pKa = 3.96TQEE7 pKa = 3.77WEE9 pKa = 4.88IYY11 pKa = 8.98PCWHH15 pKa = 6.74GKK17 pKa = 8.24PPKK20 pKa = 9.41TSGPTVVDD28 pKa = 3.35SKK30 pKa = 11.98GNIDD34 pKa = 4.04VSAIPCEE41 pKa = 4.33TEE43 pKa = 3.88SNAGVTKK50 pKa = 10.45GNHH53 pKa = 5.18VDD55 pKa = 3.94ALPYY59 pKa = 9.77WSDD62 pKa = 3.21RR63 pKa = 11.84LLVTVEE69 pKa = 3.58PRR71 pKa = 11.84VRR73 pKa = 11.84NISYY77 pKa = 10.01GVAVTRR83 pKa = 11.84MPVPPPEE90 pKa = 4.16NTCIPDD96 pKa = 3.79GDD98 pKa = 4.14PGGSKK103 pKa = 9.61TIVRR107 pKa = 11.84NKK109 pKa = 10.27YY110 pKa = 10.7SIDD113 pKa = 3.53PFEE116 pKa = 4.18WAAAPTFDD124 pKa = 5.01LPEE127 pKa = 3.94VSPAEE132 pKa = 4.06LEE134 pKa = 4.36FLRR137 pKa = 11.84GMADD141 pKa = 3.14TRR143 pKa = 11.84ALASLNKK150 pKa = 10.49AMVNLPMLFKK160 pKa = 10.55EE161 pKa = 4.09RR162 pKa = 11.84RR163 pKa = 11.84EE164 pKa = 4.14TLKK167 pKa = 10.29MAAGKK172 pKa = 10.66VGVLADD178 pKa = 3.87AARR181 pKa = 11.84SIQQRR186 pKa = 11.84DD187 pKa = 3.63FSRR190 pKa = 11.84YY191 pKa = 8.34RR192 pKa = 11.84KK193 pKa = 8.54VAKK196 pKa = 9.6RR197 pKa = 11.84DD198 pKa = 3.29RR199 pKa = 11.84RR200 pKa = 11.84RR201 pKa = 11.84VARR204 pKa = 11.84DD205 pKa = 2.92IANEE209 pKa = 3.68HH210 pKa = 6.78LEE212 pKa = 5.36FIFGWLPLIAEE223 pKa = 4.58VEE225 pKa = 4.18GAMDD229 pKa = 4.05YY230 pKa = 10.52IAQEE234 pKa = 3.72RR235 pKa = 11.84FEE237 pKa = 4.43FIKK240 pKa = 10.96GRR242 pKa = 11.84GSHH245 pKa = 6.95VITRR249 pKa = 11.84DD250 pKa = 3.38QKK252 pKa = 8.8VHH254 pKa = 5.41VSAPVLGLDD263 pKa = 3.13GWYY266 pKa = 10.56GYY268 pKa = 9.95NRR270 pKa = 11.84EE271 pKa = 3.97VARR274 pKa = 11.84AEE276 pKa = 4.04LNGIRR281 pKa = 11.84HH282 pKa = 5.84EE283 pKa = 4.66LIGMRR288 pKa = 11.84TNLRR292 pKa = 11.84MDD294 pKa = 3.38ITSAIAGDD302 pKa = 3.58ARR304 pKa = 11.84QLGFEE309 pKa = 5.15PISTLYY315 pKa = 11.42DD316 pKa = 3.44MVPLSFVSGWVTNFDD331 pKa = 3.47AYY333 pKa = 11.24VRR335 pKa = 11.84TIAPLVGLSFRR346 pKa = 11.84TGSRR350 pKa = 11.84NLRR353 pKa = 11.84KK354 pKa = 9.48QVVLDD359 pKa = 3.28VAGRR363 pKa = 11.84VLEE366 pKa = 4.34QNPPTYY372 pKa = 9.54PAPWKK377 pKa = 10.3AWVEE381 pKa = 3.85EE382 pKa = 4.05RR383 pKa = 11.84DD384 pKa = 3.55RR385 pKa = 11.84SVSLLTGNRR394 pKa = 11.84TRR396 pKa = 11.84DD397 pKa = 3.39EE398 pKa = 4.24RR399 pKa = 11.84SLVLEE404 pKa = 4.29LPKK407 pKa = 10.8ASLHH411 pKa = 5.95WDD413 pKa = 2.94VDD415 pKa = 3.73VGFNEE420 pKa = 3.83VTAAISLLIQRR431 pKa = 11.84KK432 pKa = 8.8LKK434 pKa = 9.89PLQRR438 pKa = 11.84AIGIKK443 pKa = 7.13QFRR446 pKa = 11.84YY447 pKa = 9.98RR448 pKa = 11.84GPKK451 pKa = 9.2PKK453 pKa = 9.19WLQEE457 pKa = 3.26IRR459 pKa = 11.84YY460 pKa = 8.32RR461 pKa = 11.84KK462 pKa = 8.87PP463 pKa = 2.82
MM1 pKa = 7.95RR2 pKa = 11.84SEE4 pKa = 3.96TQEE7 pKa = 3.77WEE9 pKa = 4.88IYY11 pKa = 8.98PCWHH15 pKa = 6.74GKK17 pKa = 8.24PPKK20 pKa = 9.41TSGPTVVDD28 pKa = 3.35SKK30 pKa = 11.98GNIDD34 pKa = 4.04VSAIPCEE41 pKa = 4.33TEE43 pKa = 3.88SNAGVTKK50 pKa = 10.45GNHH53 pKa = 5.18VDD55 pKa = 3.94ALPYY59 pKa = 9.77WSDD62 pKa = 3.21RR63 pKa = 11.84LLVTVEE69 pKa = 3.58PRR71 pKa = 11.84VRR73 pKa = 11.84NISYY77 pKa = 10.01GVAVTRR83 pKa = 11.84MPVPPPEE90 pKa = 4.16NTCIPDD96 pKa = 3.79GDD98 pKa = 4.14PGGSKK103 pKa = 9.61TIVRR107 pKa = 11.84NKK109 pKa = 10.27YY110 pKa = 10.7SIDD113 pKa = 3.53PFEE116 pKa = 4.18WAAAPTFDD124 pKa = 5.01LPEE127 pKa = 3.94VSPAEE132 pKa = 4.06LEE134 pKa = 4.36FLRR137 pKa = 11.84GMADD141 pKa = 3.14TRR143 pKa = 11.84ALASLNKK150 pKa = 10.49AMVNLPMLFKK160 pKa = 10.55EE161 pKa = 4.09RR162 pKa = 11.84RR163 pKa = 11.84EE164 pKa = 4.14TLKK167 pKa = 10.29MAAGKK172 pKa = 10.66VGVLADD178 pKa = 3.87AARR181 pKa = 11.84SIQQRR186 pKa = 11.84DD187 pKa = 3.63FSRR190 pKa = 11.84YY191 pKa = 8.34RR192 pKa = 11.84KK193 pKa = 8.54VAKK196 pKa = 9.6RR197 pKa = 11.84DD198 pKa = 3.29RR199 pKa = 11.84RR200 pKa = 11.84RR201 pKa = 11.84VARR204 pKa = 11.84DD205 pKa = 2.92IANEE209 pKa = 3.68HH210 pKa = 6.78LEE212 pKa = 5.36FIFGWLPLIAEE223 pKa = 4.58VEE225 pKa = 4.18GAMDD229 pKa = 4.05YY230 pKa = 10.52IAQEE234 pKa = 3.72RR235 pKa = 11.84FEE237 pKa = 4.43FIKK240 pKa = 10.96GRR242 pKa = 11.84GSHH245 pKa = 6.95VITRR249 pKa = 11.84DD250 pKa = 3.38QKK252 pKa = 8.8VHH254 pKa = 5.41VSAPVLGLDD263 pKa = 3.13GWYY266 pKa = 10.56GYY268 pKa = 9.95NRR270 pKa = 11.84EE271 pKa = 3.97VARR274 pKa = 11.84AEE276 pKa = 4.04LNGIRR281 pKa = 11.84HH282 pKa = 5.84EE283 pKa = 4.66LIGMRR288 pKa = 11.84TNLRR292 pKa = 11.84MDD294 pKa = 3.38ITSAIAGDD302 pKa = 3.58ARR304 pKa = 11.84QLGFEE309 pKa = 5.15PISTLYY315 pKa = 11.42DD316 pKa = 3.44MVPLSFVSGWVTNFDD331 pKa = 3.47AYY333 pKa = 11.24VRR335 pKa = 11.84TIAPLVGLSFRR346 pKa = 11.84TGSRR350 pKa = 11.84NLRR353 pKa = 11.84KK354 pKa = 9.48QVVLDD359 pKa = 3.28VAGRR363 pKa = 11.84VLEE366 pKa = 4.34QNPPTYY372 pKa = 9.54PAPWKK377 pKa = 10.3AWVEE381 pKa = 3.85EE382 pKa = 4.05RR383 pKa = 11.84DD384 pKa = 3.55RR385 pKa = 11.84SVSLLTGNRR394 pKa = 11.84TRR396 pKa = 11.84DD397 pKa = 3.39EE398 pKa = 4.24RR399 pKa = 11.84SLVLEE404 pKa = 4.29LPKK407 pKa = 10.8ASLHH411 pKa = 5.95WDD413 pKa = 2.94VDD415 pKa = 3.73VGFNEE420 pKa = 3.83VTAAISLLIQRR431 pKa = 11.84KK432 pKa = 8.8LKK434 pKa = 9.89PLQRR438 pKa = 11.84AIGIKK443 pKa = 7.13QFRR446 pKa = 11.84YY447 pKa = 9.98RR448 pKa = 11.84GPKK451 pKa = 9.2PKK453 pKa = 9.19WLQEE457 pKa = 3.26IRR459 pKa = 11.84YY460 pKa = 8.32RR461 pKa = 11.84KK462 pKa = 8.87PP463 pKa = 2.82
Molecular weight: 52.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1136 |
129 |
544 |
378.7 |
42.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.891 ± 0.534 | 1.32 ± 0.611 |
5.722 ± 0.388 | 6.162 ± 0.747 |
3.609 ± 0.833 | 6.074 ± 0.504 |
1.408 ± 0.154 | 4.665 ± 0.542 |
4.137 ± 0.482 | 9.243 ± 0.468 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.025 ± 0.122 | 3.257 ± 0.24 |
6.338 ± 0.789 | 2.377 ± 0.448 |
9.155 ± 0.719 | 6.954 ± 1.535 |
5.81 ± 1.045 | 7.658 ± 1.257 |
2.025 ± 0.32 | 3.169 ± 0.721 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
