
Mucilaginibacter paludis DSM 18603
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Sphingobacteriia; Sphingobacteriales; Sphingobacteriaceae; Mucilaginibacter; Mucilaginibacter paludis
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
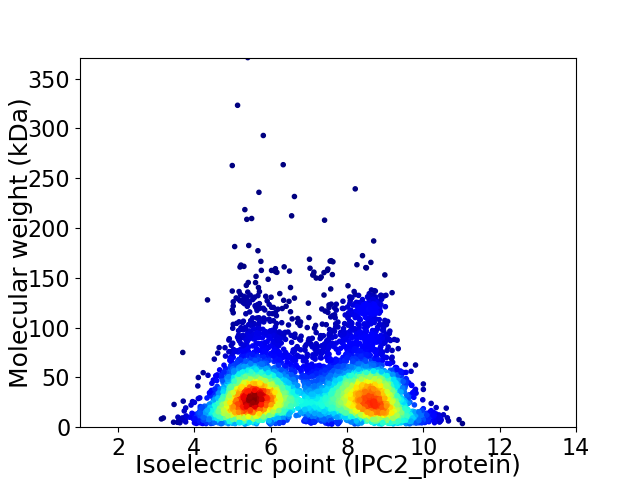
Virtual 2D-PAGE plot for 6864 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H1YIE7|H1YIE7_9SPHI Lipid A biosynthesis acyltransferase OS=Mucilaginibacter paludis DSM 18603 OX=714943 GN=Mucpa_3461 PE=4 SV=1
MM1 pKa = 7.1KK2 pKa = 10.64QNILEE7 pKa = 4.22LAGDD11 pKa = 4.39FKK13 pKa = 11.38PIGTDD18 pKa = 2.63ILRR21 pKa = 11.84GLKK24 pKa = 10.07GGNIPALTQAQLEE37 pKa = 4.36KK38 pKa = 10.93LFDD41 pKa = 3.78YY42 pKa = 11.1SGLGGFNSDD51 pKa = 2.78QSKK54 pKa = 10.58RR55 pKa = 11.84FTDD58 pKa = 3.94QIQKK62 pKa = 9.04MEE64 pKa = 4.06QTDD67 pKa = 4.29FGAGMLGAMLTTGGAPIKK85 pKa = 9.77LTEE88 pKa = 4.48QIPTQANPNLGNEE101 pKa = 4.22AAGSYY106 pKa = 10.4FPTINTLWLNPGTIDD121 pKa = 3.38VTVNSSLQSAALLDD135 pKa = 4.54TIAHH139 pKa = 6.2EE140 pKa = 5.27LYY142 pKa = 10.52HH143 pKa = 6.97SYY145 pKa = 8.37QTNILGKK152 pKa = 10.1LLGVFDD158 pKa = 4.83PNSVKK163 pKa = 10.59RR164 pKa = 11.84EE165 pKa = 3.87LDD167 pKa = 3.24ADD169 pKa = 3.99LAGAKK174 pKa = 9.3ILYY177 pKa = 10.04EE178 pKa = 3.98YY179 pKa = 10.44DD180 pKa = 3.44VKK182 pKa = 11.31FNTSYY187 pKa = 10.39YY188 pKa = 7.78NQYY191 pKa = 10.06YY192 pKa = 8.53VQEE195 pKa = 4.88HH196 pKa = 5.64YY197 pKa = 11.12SGTYY201 pKa = 7.77WQLGDD206 pKa = 4.22NNGGLYY212 pKa = 10.55GLSNITSPGVTNDD225 pKa = 3.29FGAIFDD231 pKa = 3.63QVVRR235 pKa = 11.84YY236 pKa = 9.94DD237 pKa = 4.5HH238 pKa = 7.21FNLDD242 pKa = 3.33NYY244 pKa = 11.2NYY246 pKa = 10.39LISNFLHH253 pKa = 7.06SFEE256 pKa = 4.92TPNNSYY262 pKa = 11.26NSLSAAGISDD272 pKa = 3.68TDD274 pKa = 5.36DD275 pKa = 3.41INQSAINRR283 pKa = 11.84LLFYY287 pKa = 10.64RR288 pKa = 11.84PSLLPNAPTSYY299 pKa = 9.53GTIHH303 pKa = 7.15SYY305 pKa = 11.77NDD307 pKa = 2.96VTNYY311 pKa = 8.96IQSVLNYY318 pKa = 9.93YY319 pKa = 9.04YY320 pKa = 10.86KK321 pKa = 10.75KK322 pKa = 10.6DD323 pKa = 3.68VNNGNGFGNNATITTSWHH341 pKa = 5.22YY342 pKa = 10.94SQLLGVFQNASGDD355 pKa = 3.71TMGALQIPPQVMQILVDD372 pKa = 3.76ISIEE376 pKa = 4.11SPMEE380 pKa = 3.42ITQQQFLALGAAMQNGVNLPEE401 pKa = 4.17STVGILSPPTLPPWVDD417 pKa = 3.51PSSSSANQQGLNGWYY432 pKa = 9.85YY433 pKa = 11.15DD434 pKa = 3.72SQSNSWISTIGSYY447 pKa = 10.55PITVGGGSGYY457 pKa = 10.69NFGSQDD463 pKa = 3.21PSIIGGGGVSIWYY476 pKa = 9.73NPTTDD481 pKa = 3.1HH482 pKa = 5.84TQAIASGQQPSAGYY496 pKa = 5.66TTLVKK501 pKa = 10.48SDD503 pKa = 3.68DD504 pKa = 3.73AGITKK509 pKa = 10.06AYY511 pKa = 10.17NNAYY515 pKa = 10.12DD516 pKa = 3.98YY517 pKa = 11.74NNNLPLVARR526 pKa = 11.84EE527 pKa = 3.98AAANAAALAYY537 pKa = 10.43QNAEE541 pKa = 4.08HH542 pKa = 6.74NGLSVAQANANASIAAQQAALNTLLYY568 pKa = 10.25YY569 pKa = 10.75GYY571 pKa = 10.79DD572 pKa = 3.57VDD574 pKa = 6.4DD575 pKa = 5.9LGDD578 pKa = 5.6DD579 pKa = 5.3DD580 pKa = 6.76DD581 pKa = 7.45LDD583 pKa = 6.34DD584 pKa = 6.04DD585 pKa = 5.66LDD587 pKa = 5.84DD588 pKa = 5.8IFDD591 pKa = 4.54IDD593 pKa = 6.0DD594 pKa = 5.28DD595 pKa = 6.2DD596 pKa = 5.8ILDD599 pKa = 4.18DD600 pKa = 3.81TTGTVAVSGSGNSGGGTGGSGGGSTGGGGSSGGGSSSGDD639 pKa = 3.29DD640 pKa = 3.31GSYY643 pKa = 11.26DD644 pKa = 3.84RR645 pKa = 11.84DD646 pKa = 3.37EE647 pKa = 5.66DD648 pKa = 4.41DD649 pKa = 5.64PVDD652 pKa = 3.58TSYY655 pKa = 10.18DD656 pKa = 3.24TSYY659 pKa = 11.56DD660 pKa = 3.74DD661 pKa = 5.19PGDD664 pKa = 3.95GGDD667 pKa = 4.42PSGGDD672 pKa = 3.52YY673 pKa = 11.29NGGGDD678 pKa = 3.96SVGTIYY684 pKa = 10.34IDD686 pKa = 3.45FNSYY690 pKa = 11.31DD691 pKa = 3.57NEE693 pKa = 4.21LEE695 pKa = 4.1QGITVANWWW704 pKa = 3.45
MM1 pKa = 7.1KK2 pKa = 10.64QNILEE7 pKa = 4.22LAGDD11 pKa = 4.39FKK13 pKa = 11.38PIGTDD18 pKa = 2.63ILRR21 pKa = 11.84GLKK24 pKa = 10.07GGNIPALTQAQLEE37 pKa = 4.36KK38 pKa = 10.93LFDD41 pKa = 3.78YY42 pKa = 11.1SGLGGFNSDD51 pKa = 2.78QSKK54 pKa = 10.58RR55 pKa = 11.84FTDD58 pKa = 3.94QIQKK62 pKa = 9.04MEE64 pKa = 4.06QTDD67 pKa = 4.29FGAGMLGAMLTTGGAPIKK85 pKa = 9.77LTEE88 pKa = 4.48QIPTQANPNLGNEE101 pKa = 4.22AAGSYY106 pKa = 10.4FPTINTLWLNPGTIDD121 pKa = 3.38VTVNSSLQSAALLDD135 pKa = 4.54TIAHH139 pKa = 6.2EE140 pKa = 5.27LYY142 pKa = 10.52HH143 pKa = 6.97SYY145 pKa = 8.37QTNILGKK152 pKa = 10.1LLGVFDD158 pKa = 4.83PNSVKK163 pKa = 10.59RR164 pKa = 11.84EE165 pKa = 3.87LDD167 pKa = 3.24ADD169 pKa = 3.99LAGAKK174 pKa = 9.3ILYY177 pKa = 10.04EE178 pKa = 3.98YY179 pKa = 10.44DD180 pKa = 3.44VKK182 pKa = 11.31FNTSYY187 pKa = 10.39YY188 pKa = 7.78NQYY191 pKa = 10.06YY192 pKa = 8.53VQEE195 pKa = 4.88HH196 pKa = 5.64YY197 pKa = 11.12SGTYY201 pKa = 7.77WQLGDD206 pKa = 4.22NNGGLYY212 pKa = 10.55GLSNITSPGVTNDD225 pKa = 3.29FGAIFDD231 pKa = 3.63QVVRR235 pKa = 11.84YY236 pKa = 9.94DD237 pKa = 4.5HH238 pKa = 7.21FNLDD242 pKa = 3.33NYY244 pKa = 11.2NYY246 pKa = 10.39LISNFLHH253 pKa = 7.06SFEE256 pKa = 4.92TPNNSYY262 pKa = 11.26NSLSAAGISDD272 pKa = 3.68TDD274 pKa = 5.36DD275 pKa = 3.41INQSAINRR283 pKa = 11.84LLFYY287 pKa = 10.64RR288 pKa = 11.84PSLLPNAPTSYY299 pKa = 9.53GTIHH303 pKa = 7.15SYY305 pKa = 11.77NDD307 pKa = 2.96VTNYY311 pKa = 8.96IQSVLNYY318 pKa = 9.93YY319 pKa = 9.04YY320 pKa = 10.86KK321 pKa = 10.75KK322 pKa = 10.6DD323 pKa = 3.68VNNGNGFGNNATITTSWHH341 pKa = 5.22YY342 pKa = 10.94SQLLGVFQNASGDD355 pKa = 3.71TMGALQIPPQVMQILVDD372 pKa = 3.76ISIEE376 pKa = 4.11SPMEE380 pKa = 3.42ITQQQFLALGAAMQNGVNLPEE401 pKa = 4.17STVGILSPPTLPPWVDD417 pKa = 3.51PSSSSANQQGLNGWYY432 pKa = 9.85YY433 pKa = 11.15DD434 pKa = 3.72SQSNSWISTIGSYY447 pKa = 10.55PITVGGGSGYY457 pKa = 10.69NFGSQDD463 pKa = 3.21PSIIGGGGVSIWYY476 pKa = 9.73NPTTDD481 pKa = 3.1HH482 pKa = 5.84TQAIASGQQPSAGYY496 pKa = 5.66TTLVKK501 pKa = 10.48SDD503 pKa = 3.68DD504 pKa = 3.73AGITKK509 pKa = 10.06AYY511 pKa = 10.17NNAYY515 pKa = 10.12DD516 pKa = 3.98YY517 pKa = 11.74NNNLPLVARR526 pKa = 11.84EE527 pKa = 3.98AAANAAALAYY537 pKa = 10.43QNAEE541 pKa = 4.08HH542 pKa = 6.74NGLSVAQANANASIAAQQAALNTLLYY568 pKa = 10.25YY569 pKa = 10.75GYY571 pKa = 10.79DD572 pKa = 3.57VDD574 pKa = 6.4DD575 pKa = 5.9LGDD578 pKa = 5.6DD579 pKa = 5.3DD580 pKa = 6.76DD581 pKa = 7.45LDD583 pKa = 6.34DD584 pKa = 6.04DD585 pKa = 5.66LDD587 pKa = 5.84DD588 pKa = 5.8IFDD591 pKa = 4.54IDD593 pKa = 6.0DD594 pKa = 5.28DD595 pKa = 6.2DD596 pKa = 5.8ILDD599 pKa = 4.18DD600 pKa = 3.81TTGTVAVSGSGNSGGGTGGSGGGSTGGGGSSGGGSSSGDD639 pKa = 3.29DD640 pKa = 3.31GSYY643 pKa = 11.26DD644 pKa = 3.84RR645 pKa = 11.84DD646 pKa = 3.37EE647 pKa = 5.66DD648 pKa = 4.41DD649 pKa = 5.64PVDD652 pKa = 3.58TSYY655 pKa = 10.18DD656 pKa = 3.24TSYY659 pKa = 11.56DD660 pKa = 3.74DD661 pKa = 5.19PGDD664 pKa = 3.95GGDD667 pKa = 4.42PSGGDD672 pKa = 3.52YY673 pKa = 11.29NGGGDD678 pKa = 3.96SVGTIYY684 pKa = 10.34IDD686 pKa = 3.45FNSYY690 pKa = 11.31DD691 pKa = 3.57NEE693 pKa = 4.21LEE695 pKa = 4.1QGITVANWWW704 pKa = 3.45
Molecular weight: 75.07 kDa
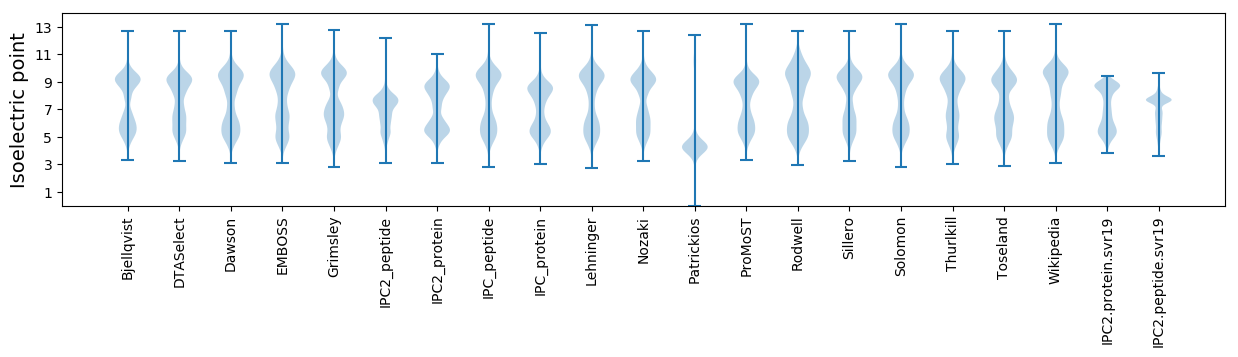
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H1XZP1|H1XZP1_9SPHI NAD-dependent epimerase/dehydratase OS=Mucilaginibacter paludis DSM 18603 OX=714943 GN=Mucpa_3635 PE=4 SV=1
MM1 pKa = 7.84PSGKK5 pKa = 9.39KK6 pKa = 9.62RR7 pKa = 11.84KK8 pKa = 6.72RR9 pKa = 11.84HH10 pKa = 5.17KK11 pKa = 10.05MATHH15 pKa = 6.02KK16 pKa = 10.34RR17 pKa = 11.84KK18 pKa = 9.84KK19 pKa = 9.28RR20 pKa = 11.84LRR22 pKa = 11.84KK23 pKa = 9.26NRR25 pKa = 11.84HH26 pKa = 4.71KK27 pKa = 10.87KK28 pKa = 9.36KK29 pKa = 10.77
MM1 pKa = 7.84PSGKK5 pKa = 9.39KK6 pKa = 9.62RR7 pKa = 11.84KK8 pKa = 6.72RR9 pKa = 11.84HH10 pKa = 5.17KK11 pKa = 10.05MATHH15 pKa = 6.02KK16 pKa = 10.34RR17 pKa = 11.84KK18 pKa = 9.84KK19 pKa = 9.28RR20 pKa = 11.84LRR22 pKa = 11.84KK23 pKa = 9.26NRR25 pKa = 11.84HH26 pKa = 4.71KK27 pKa = 10.87KK28 pKa = 9.36KK29 pKa = 10.77
Molecular weight: 3.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2350594 |
29 |
3731 |
342.5 |
38.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.605 ± 0.028 | 0.807 ± 0.009 |
5.383 ± 0.018 | 5.107 ± 0.031 |
4.773 ± 0.02 | 6.709 ± 0.03 |
1.903 ± 0.014 | 7.21 ± 0.023 |
6.849 ± 0.028 | 9.584 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.224 ± 0.014 | 5.817 ± 0.028 |
3.903 ± 0.015 | 4.033 ± 0.02 |
3.902 ± 0.016 | 6.446 ± 0.025 |
6.006 ± 0.033 | 6.266 ± 0.02 |
1.166 ± 0.011 | 4.307 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
