
Murid herpesvirus 1 (strain Smith) (MuHV-1) (Mouse cytomegalovirus)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Betaherpesvirinae; Muromegalovirus; Murid betaherpesvirus 1
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
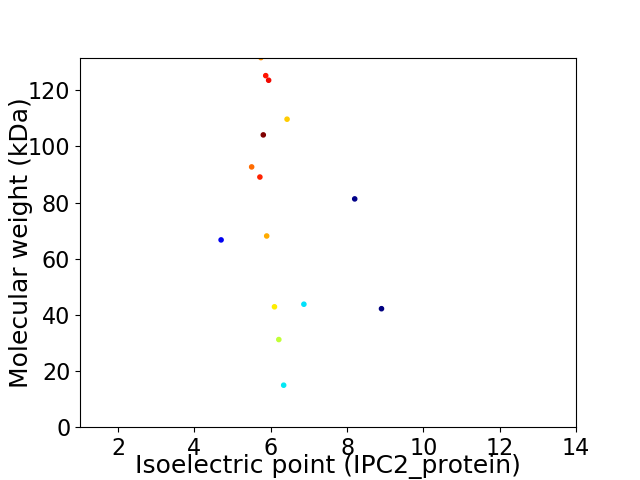
Virtual 2D-PAGE plot for 15 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P24909|VIE2_MUHVS Immediate-early protein 2 OS=Murid herpesvirus 1 (strain Smith) OX=10367 GN=IE2 PE=3 SV=1
MM1 pKa = 7.37EE2 pKa = 5.37PAAPSCNMIMIADD15 pKa = 3.82QASVNAHH22 pKa = 5.32GRR24 pKa = 11.84HH25 pKa = 5.59LDD27 pKa = 3.48EE28 pKa = 4.46NRR30 pKa = 11.84VYY32 pKa = 10.74PSDD35 pKa = 3.65KK36 pKa = 10.84VPAHH40 pKa = 5.79VANKK44 pKa = 9.38ILEE47 pKa = 4.64SGTEE51 pKa = 4.03TVRR54 pKa = 11.84CDD56 pKa = 3.37LTLEE60 pKa = 4.51DD61 pKa = 3.93MLGDD65 pKa = 3.92YY66 pKa = 10.52EE67 pKa = 4.94YY68 pKa = 11.12DD69 pKa = 3.87DD70 pKa = 3.83PTEE73 pKa = 4.03EE74 pKa = 4.43EE75 pKa = 4.78KK76 pKa = 10.87ILMDD80 pKa = 5.63RR81 pKa = 11.84IADD84 pKa = 3.53HH85 pKa = 6.57VGNDD89 pKa = 3.48NSDD92 pKa = 3.36MAIKK96 pKa = 9.79HH97 pKa = 5.19AAVRR101 pKa = 11.84SVLLSCKK108 pKa = 9.47IAHH111 pKa = 7.08LMIKK115 pKa = 10.05QNYY118 pKa = 7.79QSAINSATNILCQLANDD135 pKa = 3.06IFEE138 pKa = 4.79RR139 pKa = 11.84IEE141 pKa = 3.89RR142 pKa = 11.84QRR144 pKa = 11.84KK145 pKa = 7.81MIYY148 pKa = 9.69GCFRR152 pKa = 11.84SEE154 pKa = 4.0FDD156 pKa = 3.25NVQLGRR162 pKa = 11.84LMYY165 pKa = 10.99DD166 pKa = 3.69MYY168 pKa = 11.16PHH170 pKa = 7.14FMPTNLGPSEE180 pKa = 3.96KK181 pKa = 9.44RR182 pKa = 11.84VWMSYY187 pKa = 9.45VGEE190 pKa = 4.71AIVAATNIDD199 pKa = 3.43HH200 pKa = 7.3ALDD203 pKa = 4.09EE204 pKa = 4.33RR205 pKa = 11.84AAWAKK210 pKa = 9.43TDD212 pKa = 3.51CSLPGEE218 pKa = 4.97FKK220 pKa = 10.89PEE222 pKa = 3.85LCVLVGAIRR231 pKa = 11.84RR232 pKa = 11.84LHH234 pKa = 7.2DD235 pKa = 4.02PPCYY239 pKa = 9.78TKK241 pKa = 10.67PFLDD245 pKa = 4.35AKK247 pKa = 10.41SQLAVWQQMKK257 pKa = 10.45AIEE260 pKa = 4.33SEE262 pKa = 4.57SVSTHH267 pKa = 3.93VVVVEE272 pKa = 3.74ALKK275 pKa = 10.88LRR277 pKa = 11.84EE278 pKa = 4.06NLAKK282 pKa = 10.34AVQEE286 pKa = 4.18TIAYY290 pKa = 7.65EE291 pKa = 3.52RR292 pKa = 11.84HH293 pKa = 5.14QYY295 pKa = 10.43HH296 pKa = 6.4RR297 pKa = 11.84VCQMMCNNMKK307 pKa = 10.62DD308 pKa = 3.75HH309 pKa = 7.58LEE311 pKa = 4.33TTCMLARR318 pKa = 11.84GRR320 pKa = 11.84TLATLADD327 pKa = 4.06LRR329 pKa = 11.84STRR332 pKa = 11.84YY333 pKa = 9.95NLALFLLSEE342 pKa = 3.89MHH344 pKa = 6.5IFDD347 pKa = 4.66SFTMPRR353 pKa = 11.84IRR355 pKa = 11.84GAMKK359 pKa = 9.74QARR362 pKa = 11.84CMSYY366 pKa = 9.98VEE368 pKa = 4.22RR369 pKa = 11.84TISLAKK375 pKa = 10.1FRR377 pKa = 11.84EE378 pKa = 4.2LADD381 pKa = 3.61RR382 pKa = 11.84VHH384 pKa = 6.21NRR386 pKa = 11.84SAPSPQGVIEE396 pKa = 4.2EE397 pKa = 4.03QQQAGEE403 pKa = 4.48EE404 pKa = 4.2EE405 pKa = 4.16QQQQQEE411 pKa = 4.09IEE413 pKa = 4.51YY414 pKa = 9.66DD415 pKa = 3.49PEE417 pKa = 4.08MPPLEE422 pKa = 4.27RR423 pKa = 11.84EE424 pKa = 4.14EE425 pKa = 4.13EE426 pKa = 4.21QEE428 pKa = 3.95DD429 pKa = 4.0EE430 pKa = 4.42QVEE433 pKa = 4.38EE434 pKa = 4.54EE435 pKa = 4.37PPADD439 pKa = 3.91EE440 pKa = 4.69EE441 pKa = 4.19EE442 pKa = 4.64GGAVGGVTQEE452 pKa = 4.18EE453 pKa = 4.64PAGEE457 pKa = 4.01ATEE460 pKa = 4.04EE461 pKa = 4.19AEE463 pKa = 4.45EE464 pKa = 5.27DD465 pKa = 3.65EE466 pKa = 4.69SQPGPSDD473 pKa = 3.4NQVVPEE479 pKa = 4.13SSEE482 pKa = 4.27TPTPAEE488 pKa = 4.23DD489 pKa = 4.9EE490 pKa = 4.43EE491 pKa = 4.63TQSADD496 pKa = 3.16EE497 pKa = 4.92GEE499 pKa = 4.37SQEE502 pKa = 5.53LEE504 pKa = 4.19GSQQLILSRR513 pKa = 11.84PAAPLTDD520 pKa = 4.0SEE522 pKa = 4.56TDD524 pKa = 3.27SDD526 pKa = 5.25SEE528 pKa = 5.18DD529 pKa = 3.8DD530 pKa = 5.11DD531 pKa = 4.04EE532 pKa = 4.61VTRR535 pKa = 11.84IPVGFSLMTSPVLQPTTRR553 pKa = 11.84SATAAASSGTAPRR566 pKa = 11.84PALKK570 pKa = 9.96RR571 pKa = 11.84QYY573 pKa = 11.74AMVHH577 pKa = 5.0TRR579 pKa = 11.84SKK581 pKa = 10.79SSEE584 pKa = 3.77NQQQPKK590 pKa = 10.07KK591 pKa = 9.94KK592 pKa = 9.98SKK594 pKa = 10.33KK595 pKa = 9.54
MM1 pKa = 7.37EE2 pKa = 5.37PAAPSCNMIMIADD15 pKa = 3.82QASVNAHH22 pKa = 5.32GRR24 pKa = 11.84HH25 pKa = 5.59LDD27 pKa = 3.48EE28 pKa = 4.46NRR30 pKa = 11.84VYY32 pKa = 10.74PSDD35 pKa = 3.65KK36 pKa = 10.84VPAHH40 pKa = 5.79VANKK44 pKa = 9.38ILEE47 pKa = 4.64SGTEE51 pKa = 4.03TVRR54 pKa = 11.84CDD56 pKa = 3.37LTLEE60 pKa = 4.51DD61 pKa = 3.93MLGDD65 pKa = 3.92YY66 pKa = 10.52EE67 pKa = 4.94YY68 pKa = 11.12DD69 pKa = 3.87DD70 pKa = 3.83PTEE73 pKa = 4.03EE74 pKa = 4.43EE75 pKa = 4.78KK76 pKa = 10.87ILMDD80 pKa = 5.63RR81 pKa = 11.84IADD84 pKa = 3.53HH85 pKa = 6.57VGNDD89 pKa = 3.48NSDD92 pKa = 3.36MAIKK96 pKa = 9.79HH97 pKa = 5.19AAVRR101 pKa = 11.84SVLLSCKK108 pKa = 9.47IAHH111 pKa = 7.08LMIKK115 pKa = 10.05QNYY118 pKa = 7.79QSAINSATNILCQLANDD135 pKa = 3.06IFEE138 pKa = 4.79RR139 pKa = 11.84IEE141 pKa = 3.89RR142 pKa = 11.84QRR144 pKa = 11.84KK145 pKa = 7.81MIYY148 pKa = 9.69GCFRR152 pKa = 11.84SEE154 pKa = 4.0FDD156 pKa = 3.25NVQLGRR162 pKa = 11.84LMYY165 pKa = 10.99DD166 pKa = 3.69MYY168 pKa = 11.16PHH170 pKa = 7.14FMPTNLGPSEE180 pKa = 3.96KK181 pKa = 9.44RR182 pKa = 11.84VWMSYY187 pKa = 9.45VGEE190 pKa = 4.71AIVAATNIDD199 pKa = 3.43HH200 pKa = 7.3ALDD203 pKa = 4.09EE204 pKa = 4.33RR205 pKa = 11.84AAWAKK210 pKa = 9.43TDD212 pKa = 3.51CSLPGEE218 pKa = 4.97FKK220 pKa = 10.89PEE222 pKa = 3.85LCVLVGAIRR231 pKa = 11.84RR232 pKa = 11.84LHH234 pKa = 7.2DD235 pKa = 4.02PPCYY239 pKa = 9.78TKK241 pKa = 10.67PFLDD245 pKa = 4.35AKK247 pKa = 10.41SQLAVWQQMKK257 pKa = 10.45AIEE260 pKa = 4.33SEE262 pKa = 4.57SVSTHH267 pKa = 3.93VVVVEE272 pKa = 3.74ALKK275 pKa = 10.88LRR277 pKa = 11.84EE278 pKa = 4.06NLAKK282 pKa = 10.34AVQEE286 pKa = 4.18TIAYY290 pKa = 7.65EE291 pKa = 3.52RR292 pKa = 11.84HH293 pKa = 5.14QYY295 pKa = 10.43HH296 pKa = 6.4RR297 pKa = 11.84VCQMMCNNMKK307 pKa = 10.62DD308 pKa = 3.75HH309 pKa = 7.58LEE311 pKa = 4.33TTCMLARR318 pKa = 11.84GRR320 pKa = 11.84TLATLADD327 pKa = 4.06LRR329 pKa = 11.84STRR332 pKa = 11.84YY333 pKa = 9.95NLALFLLSEE342 pKa = 3.89MHH344 pKa = 6.5IFDD347 pKa = 4.66SFTMPRR353 pKa = 11.84IRR355 pKa = 11.84GAMKK359 pKa = 9.74QARR362 pKa = 11.84CMSYY366 pKa = 9.98VEE368 pKa = 4.22RR369 pKa = 11.84TISLAKK375 pKa = 10.1FRR377 pKa = 11.84EE378 pKa = 4.2LADD381 pKa = 3.61RR382 pKa = 11.84VHH384 pKa = 6.21NRR386 pKa = 11.84SAPSPQGVIEE396 pKa = 4.2EE397 pKa = 4.03QQQAGEE403 pKa = 4.48EE404 pKa = 4.2EE405 pKa = 4.16QQQQQEE411 pKa = 4.09IEE413 pKa = 4.51YY414 pKa = 9.66DD415 pKa = 3.49PEE417 pKa = 4.08MPPLEE422 pKa = 4.27RR423 pKa = 11.84EE424 pKa = 4.14EE425 pKa = 4.13EE426 pKa = 4.21QEE428 pKa = 3.95DD429 pKa = 4.0EE430 pKa = 4.42QVEE433 pKa = 4.38EE434 pKa = 4.54EE435 pKa = 4.37PPADD439 pKa = 3.91EE440 pKa = 4.69EE441 pKa = 4.19EE442 pKa = 4.64GGAVGGVTQEE452 pKa = 4.18EE453 pKa = 4.64PAGEE457 pKa = 4.01ATEE460 pKa = 4.04EE461 pKa = 4.19AEE463 pKa = 4.45EE464 pKa = 5.27DD465 pKa = 3.65EE466 pKa = 4.69SQPGPSDD473 pKa = 3.4NQVVPEE479 pKa = 4.13SSEE482 pKa = 4.27TPTPAEE488 pKa = 4.23DD489 pKa = 4.9EE490 pKa = 4.43EE491 pKa = 4.63TQSADD496 pKa = 3.16EE497 pKa = 4.92GEE499 pKa = 4.37SQEE502 pKa = 5.53LEE504 pKa = 4.19GSQQLILSRR513 pKa = 11.84PAAPLTDD520 pKa = 4.0SEE522 pKa = 4.56TDD524 pKa = 3.27SDD526 pKa = 5.25SEE528 pKa = 5.18DD529 pKa = 3.8DD530 pKa = 5.11DD531 pKa = 4.04EE532 pKa = 4.61VTRR535 pKa = 11.84IPVGFSLMTSPVLQPTTRR553 pKa = 11.84SATAAASSGTAPRR566 pKa = 11.84PALKK570 pKa = 9.96RR571 pKa = 11.84QYY573 pKa = 11.74AMVHH577 pKa = 5.0TRR579 pKa = 11.84SKK581 pKa = 10.79SSEE584 pKa = 3.77NQQQPKK590 pKa = 10.07KK591 pKa = 9.94KK592 pKa = 9.98SKK594 pKa = 10.33KK595 pKa = 9.54
Molecular weight: 66.71 kDa
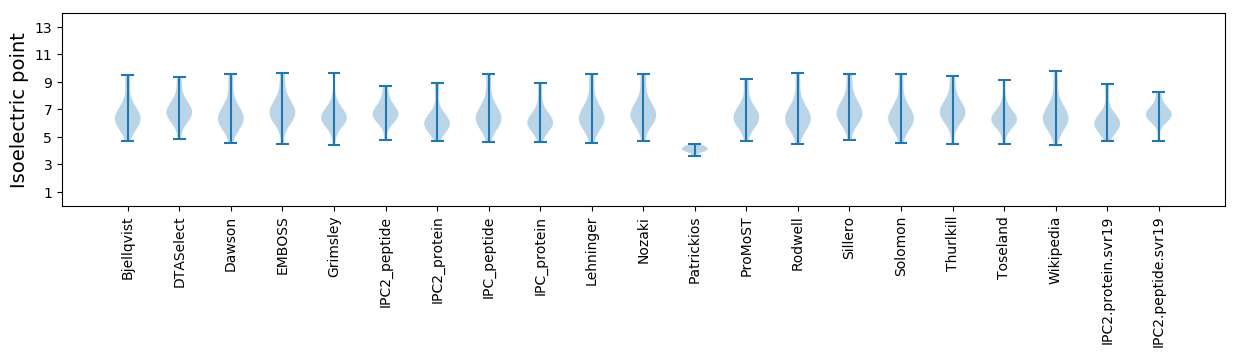
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q83207|UL33_MUHVS G-protein coupled receptor homolog M33 OS=Murid herpesvirus 1 (strain Smith) OX=10367 PE=3 SV=1
MM1 pKa = 7.91DD2 pKa = 3.75VLLGRR7 pKa = 11.84DD8 pKa = 3.39EE9 pKa = 4.59TMDD12 pKa = 3.42EE13 pKa = 3.98SDD15 pKa = 3.79YY16 pKa = 11.53LHH18 pKa = 6.87VNNTCAPSLGLSIARR33 pKa = 11.84DD34 pKa = 3.42AEE36 pKa = 4.35TAINTVIILIGGPMNFVVLTTQLLSNRR63 pKa = 11.84IYY65 pKa = 10.6RR66 pKa = 11.84SSAPTLYY73 pKa = 7.84MTNLYY78 pKa = 9.43FANLLTVTMLPFLILSNRR96 pKa = 11.84GQISSSPEE104 pKa = 3.56GCKK107 pKa = 10.08LVAVTYY113 pKa = 9.57YY114 pKa = 10.86ASCTAGFSTLALISVNRR131 pKa = 11.84YY132 pKa = 8.7RR133 pKa = 11.84VIHH136 pKa = 6.05QSTNKK141 pKa = 9.66NAAGSKK147 pKa = 9.33KK148 pKa = 8.96KK149 pKa = 9.31TYY151 pKa = 10.44GVLALVWLTSLMCASPAPTYY171 pKa = 9.89VTVLAHH177 pKa = 7.55DD178 pKa = 4.55GDD180 pKa = 4.55TPDD183 pKa = 4.62SVHH186 pKa = 5.51EE187 pKa = 4.01TCIIFFNYY195 pKa = 10.2DD196 pKa = 3.04QVKK199 pKa = 8.12TVLATFKK206 pKa = 10.77ILICIVWGVMPVVMMTWFYY225 pKa = 11.29LFFYY229 pKa = 10.42KK230 pKa = 10.33RR231 pKa = 11.84LKK233 pKa = 8.24LTSYY237 pKa = 10.31RR238 pKa = 11.84RR239 pKa = 11.84RR240 pKa = 11.84SHH242 pKa = 5.87TLAFVSTLILSFLVLQTPFVGIMIFDD268 pKa = 4.24SYY270 pKa = 11.94AVIEE274 pKa = 4.1WDD276 pKa = 3.93VTCEE280 pKa = 4.3SINSRR285 pKa = 11.84DD286 pKa = 3.63AVAMLARR293 pKa = 11.84VVPNFHH299 pKa = 7.08CMLNPVLYY307 pKa = 10.44AFLGRR312 pKa = 11.84DD313 pKa = 3.42FNKK316 pKa = 10.33RR317 pKa = 11.84FMQCITGKK325 pKa = 9.9LFSRR329 pKa = 11.84RR330 pKa = 11.84RR331 pKa = 11.84MLQEE335 pKa = 3.52RR336 pKa = 11.84AGVRR340 pKa = 11.84SPSTPHH346 pKa = 6.15RR347 pKa = 11.84AARR350 pKa = 11.84QLAKK354 pKa = 10.26IGTLTRR360 pKa = 11.84SCSRR364 pKa = 11.84SEE366 pKa = 4.07LQRR369 pKa = 11.84SASAPPPQQ377 pKa = 4.54
MM1 pKa = 7.91DD2 pKa = 3.75VLLGRR7 pKa = 11.84DD8 pKa = 3.39EE9 pKa = 4.59TMDD12 pKa = 3.42EE13 pKa = 3.98SDD15 pKa = 3.79YY16 pKa = 11.53LHH18 pKa = 6.87VNNTCAPSLGLSIARR33 pKa = 11.84DD34 pKa = 3.42AEE36 pKa = 4.35TAINTVIILIGGPMNFVVLTTQLLSNRR63 pKa = 11.84IYY65 pKa = 10.6RR66 pKa = 11.84SSAPTLYY73 pKa = 7.84MTNLYY78 pKa = 9.43FANLLTVTMLPFLILSNRR96 pKa = 11.84GQISSSPEE104 pKa = 3.56GCKK107 pKa = 10.08LVAVTYY113 pKa = 9.57YY114 pKa = 10.86ASCTAGFSTLALISVNRR131 pKa = 11.84YY132 pKa = 8.7RR133 pKa = 11.84VIHH136 pKa = 6.05QSTNKK141 pKa = 9.66NAAGSKK147 pKa = 9.33KK148 pKa = 8.96KK149 pKa = 9.31TYY151 pKa = 10.44GVLALVWLTSLMCASPAPTYY171 pKa = 9.89VTVLAHH177 pKa = 7.55DD178 pKa = 4.55GDD180 pKa = 4.55TPDD183 pKa = 4.62SVHH186 pKa = 5.51EE187 pKa = 4.01TCIIFFNYY195 pKa = 10.2DD196 pKa = 3.04QVKK199 pKa = 8.12TVLATFKK206 pKa = 10.77ILICIVWGVMPVVMMTWFYY225 pKa = 11.29LFFYY229 pKa = 10.42KK230 pKa = 10.33RR231 pKa = 11.84LKK233 pKa = 8.24LTSYY237 pKa = 10.31RR238 pKa = 11.84RR239 pKa = 11.84RR240 pKa = 11.84SHH242 pKa = 5.87TLAFVSTLILSFLVLQTPFVGIMIFDD268 pKa = 4.24SYY270 pKa = 11.94AVIEE274 pKa = 4.1WDD276 pKa = 3.93VTCEE280 pKa = 4.3SINSRR285 pKa = 11.84DD286 pKa = 3.63AVAMLARR293 pKa = 11.84VVPNFHH299 pKa = 7.08CMLNPVLYY307 pKa = 10.44AFLGRR312 pKa = 11.84DD313 pKa = 3.42FNKK316 pKa = 10.33RR317 pKa = 11.84FMQCITGKK325 pKa = 9.9LFSRR329 pKa = 11.84RR330 pKa = 11.84RR331 pKa = 11.84MLQEE335 pKa = 3.52RR336 pKa = 11.84AGVRR340 pKa = 11.84SPSTPHH346 pKa = 6.15RR347 pKa = 11.84AARR350 pKa = 11.84QLAKK354 pKa = 10.26IGTLTRR360 pKa = 11.84SCSRR364 pKa = 11.84SEE366 pKa = 4.07LQRR369 pKa = 11.84SASAPPPQQ377 pKa = 4.54
Molecular weight: 42.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10487 |
139 |
1191 |
699.1 |
77.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.153 ± 0.67 | 2.393 ± 0.135 |
6.255 ± 0.263 | 6.007 ± 0.473 |
3.986 ± 0.355 | 6.17 ± 0.411 |
2.327 ± 0.249 | 4.262 ± 0.216 |
4.1 ± 0.337 | 8.592 ± 0.493 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.784 ± 0.198 | 3.805 ± 0.188 |
5.493 ± 0.428 | 3.042 ± 0.237 |
7.257 ± 0.417 | 8.191 ± 0.517 |
5.874 ± 0.465 | 7.123 ± 0.497 |
0.687 ± 0.079 | 3.5 ± 0.23 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
