
Wuhan insect virus 17
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
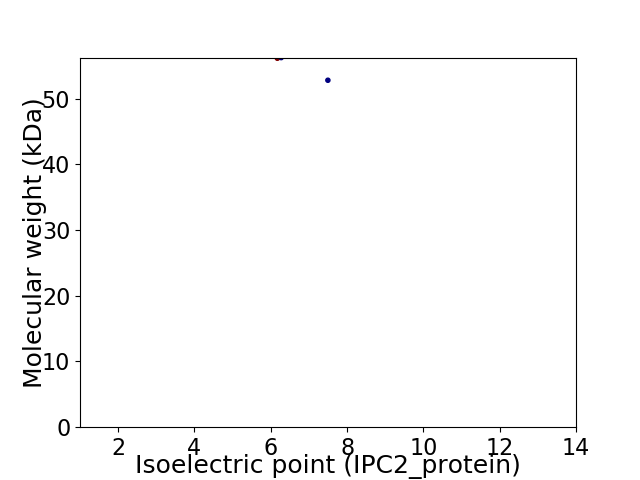
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1L3KEV4|A0A1L3KEV4_9VIRU RNA-directed RNA polymerase OS=Wuhan insect virus 17 OX=1923721 PE=4 SV=1
MM1 pKa = 8.09DD2 pKa = 6.11PLDD5 pKa = 4.34CGIMAGVAEE14 pKa = 4.83APVDD18 pKa = 3.86VQRR21 pKa = 11.84AVADD25 pKa = 3.98ALAQAIAAHH34 pKa = 5.21QLRR37 pKa = 11.84YY38 pKa = 9.56PGVLAAVAEE47 pKa = 4.34IGGCVGGVVKK57 pKa = 10.42AAAKK61 pKa = 10.05AVGGAVLSMRR71 pKa = 11.84SPTLVEE77 pKa = 4.31AGLAVALVGVGAKK90 pKa = 10.19AYY92 pKa = 10.63VPALKK97 pKa = 10.32KK98 pKa = 10.18RR99 pKa = 11.84VRR101 pKa = 11.84GWRR104 pKa = 11.84KK105 pKa = 8.08PAVKK109 pKa = 9.9EE110 pKa = 4.19PEE112 pKa = 4.29PEE114 pKa = 3.88QPPAIVPPPEE124 pKa = 3.68IAEE127 pKa = 4.3LLRR130 pKa = 11.84EE131 pKa = 4.14EE132 pKa = 4.59YY133 pKa = 10.94VPEE136 pKa = 4.05STVAGSEE143 pKa = 4.47FIPMEE148 pKa = 4.47RR149 pKa = 11.84PTHH152 pKa = 4.58QVCFGIHH159 pKa = 5.22EE160 pKa = 4.27WDD162 pKa = 3.4AKK164 pKa = 10.45LGRR167 pKa = 11.84LGFKK171 pKa = 10.84VMGAGCLVKK180 pKa = 8.88WTLGANGSAHH190 pKa = 6.44YY191 pKa = 9.88YY192 pKa = 10.12VIVHH196 pKa = 6.29EE197 pKa = 4.33HH198 pKa = 5.6TLVHH202 pKa = 6.18SSHH205 pKa = 5.98MVGHH209 pKa = 6.01NRR211 pKa = 11.84YY212 pKa = 8.19PLDD215 pKa = 3.57YY216 pKa = 10.34KK217 pKa = 11.13VEE219 pKa = 4.0LMRR222 pKa = 11.84NSLQHH227 pKa = 7.1LDD229 pKa = 4.28LKK231 pKa = 10.74CLPATVFHH239 pKa = 6.64QDD241 pKa = 2.59HH242 pKa = 6.04VAIEE246 pKa = 4.23LTQSQVSQLGLAPVRR261 pKa = 11.84RR262 pKa = 11.84IGVLDD267 pKa = 3.34KK268 pKa = 9.56RR269 pKa = 11.84TSVAIVGPNGLGSSGDD285 pKa = 3.48AEE287 pKa = 4.49MISDD291 pKa = 4.01GSIGQMSYY299 pKa = 9.85TGSTIGGFSGCAYY312 pKa = 10.18VSGSAVFGIHH322 pKa = 5.33QAGGARR328 pKa = 11.84NTGYY332 pKa = 8.69TLSYY336 pKa = 10.49LLKK339 pKa = 10.81KK340 pKa = 10.79LMLEE344 pKa = 4.15LGFCKK349 pKa = 9.96RR350 pKa = 11.84ARR352 pKa = 11.84VEE354 pKa = 3.75RR355 pKa = 11.84VAIYY359 pKa = 10.32VCGHH363 pKa = 6.64PVYY366 pKa = 10.24TPPCGDD372 pKa = 3.3LSVPEE377 pKa = 5.26SSDD380 pKa = 3.42QYY382 pKa = 11.58FKK384 pKa = 11.21DD385 pKa = 3.4LYY387 pKa = 9.48MEE389 pKa = 4.5KK390 pKa = 10.54VPIKK394 pKa = 9.71WEE396 pKa = 3.76TWGVDD401 pKa = 3.16EE402 pKa = 5.02VLIEE406 pKa = 3.78IRR408 pKa = 11.84GRR410 pKa = 11.84TSVHH414 pKa = 6.56DD415 pKa = 3.71VGTLRR420 pKa = 11.84RR421 pKa = 11.84KK422 pKa = 10.08FGRR425 pKa = 11.84AWLEE429 pKa = 3.89NLSKK433 pKa = 10.72PVGQRR438 pKa = 11.84HH439 pKa = 5.81ARR441 pKa = 11.84DD442 pKa = 3.59FDD444 pKa = 4.37DD445 pKa = 6.38SIPEE449 pKa = 4.01CAAEE453 pKa = 3.98PLEE456 pKa = 4.64IPSQSGEE463 pKa = 4.12SRR465 pKa = 11.84SPGALSEE472 pKa = 4.46SASSQGVQACQLAVLTATFATLSPEE497 pKa = 3.94LKK499 pKa = 10.54AAFLTQCAQSMTSCSSPVRR518 pKa = 11.84QEE520 pKa = 4.01TGSTQAA526 pKa = 4.29
MM1 pKa = 8.09DD2 pKa = 6.11PLDD5 pKa = 4.34CGIMAGVAEE14 pKa = 4.83APVDD18 pKa = 3.86VQRR21 pKa = 11.84AVADD25 pKa = 3.98ALAQAIAAHH34 pKa = 5.21QLRR37 pKa = 11.84YY38 pKa = 9.56PGVLAAVAEE47 pKa = 4.34IGGCVGGVVKK57 pKa = 10.42AAAKK61 pKa = 10.05AVGGAVLSMRR71 pKa = 11.84SPTLVEE77 pKa = 4.31AGLAVALVGVGAKK90 pKa = 10.19AYY92 pKa = 10.63VPALKK97 pKa = 10.32KK98 pKa = 10.18RR99 pKa = 11.84VRR101 pKa = 11.84GWRR104 pKa = 11.84KK105 pKa = 8.08PAVKK109 pKa = 9.9EE110 pKa = 4.19PEE112 pKa = 4.29PEE114 pKa = 3.88QPPAIVPPPEE124 pKa = 3.68IAEE127 pKa = 4.3LLRR130 pKa = 11.84EE131 pKa = 4.14EE132 pKa = 4.59YY133 pKa = 10.94VPEE136 pKa = 4.05STVAGSEE143 pKa = 4.47FIPMEE148 pKa = 4.47RR149 pKa = 11.84PTHH152 pKa = 4.58QVCFGIHH159 pKa = 5.22EE160 pKa = 4.27WDD162 pKa = 3.4AKK164 pKa = 10.45LGRR167 pKa = 11.84LGFKK171 pKa = 10.84VMGAGCLVKK180 pKa = 8.88WTLGANGSAHH190 pKa = 6.44YY191 pKa = 9.88YY192 pKa = 10.12VIVHH196 pKa = 6.29EE197 pKa = 4.33HH198 pKa = 5.6TLVHH202 pKa = 6.18SSHH205 pKa = 5.98MVGHH209 pKa = 6.01NRR211 pKa = 11.84YY212 pKa = 8.19PLDD215 pKa = 3.57YY216 pKa = 10.34KK217 pKa = 11.13VEE219 pKa = 4.0LMRR222 pKa = 11.84NSLQHH227 pKa = 7.1LDD229 pKa = 4.28LKK231 pKa = 10.74CLPATVFHH239 pKa = 6.64QDD241 pKa = 2.59HH242 pKa = 6.04VAIEE246 pKa = 4.23LTQSQVSQLGLAPVRR261 pKa = 11.84RR262 pKa = 11.84IGVLDD267 pKa = 3.34KK268 pKa = 9.56RR269 pKa = 11.84TSVAIVGPNGLGSSGDD285 pKa = 3.48AEE287 pKa = 4.49MISDD291 pKa = 4.01GSIGQMSYY299 pKa = 9.85TGSTIGGFSGCAYY312 pKa = 10.18VSGSAVFGIHH322 pKa = 5.33QAGGARR328 pKa = 11.84NTGYY332 pKa = 8.69TLSYY336 pKa = 10.49LLKK339 pKa = 10.81KK340 pKa = 10.79LMLEE344 pKa = 4.15LGFCKK349 pKa = 9.96RR350 pKa = 11.84ARR352 pKa = 11.84VEE354 pKa = 3.75RR355 pKa = 11.84VAIYY359 pKa = 10.32VCGHH363 pKa = 6.64PVYY366 pKa = 10.24TPPCGDD372 pKa = 3.3LSVPEE377 pKa = 5.26SSDD380 pKa = 3.42QYY382 pKa = 11.58FKK384 pKa = 11.21DD385 pKa = 3.4LYY387 pKa = 9.48MEE389 pKa = 4.5KK390 pKa = 10.54VPIKK394 pKa = 9.71WEE396 pKa = 3.76TWGVDD401 pKa = 3.16EE402 pKa = 5.02VLIEE406 pKa = 3.78IRR408 pKa = 11.84GRR410 pKa = 11.84TSVHH414 pKa = 6.56DD415 pKa = 3.71VGTLRR420 pKa = 11.84RR421 pKa = 11.84KK422 pKa = 10.08FGRR425 pKa = 11.84AWLEE429 pKa = 3.89NLSKK433 pKa = 10.72PVGQRR438 pKa = 11.84HH439 pKa = 5.81ARR441 pKa = 11.84DD442 pKa = 3.59FDD444 pKa = 4.37DD445 pKa = 6.38SIPEE449 pKa = 4.01CAAEE453 pKa = 3.98PLEE456 pKa = 4.64IPSQSGEE463 pKa = 4.12SRR465 pKa = 11.84SPGALSEE472 pKa = 4.46SASSQGVQACQLAVLTATFATLSPEE497 pKa = 3.94LKK499 pKa = 10.54AAFLTQCAQSMTSCSSPVRR518 pKa = 11.84QEE520 pKa = 4.01TGSTQAA526 pKa = 4.29
Molecular weight: 56.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEV4|A0A1L3KEV4_9VIRU RNA-directed RNA polymerase OS=Wuhan insect virus 17 OX=1923721 PE=4 SV=1
MM1 pKa = 7.86RR2 pKa = 11.84SVDD5 pKa = 4.46DD6 pKa = 4.11EE7 pKa = 4.78LLFPSEE13 pKa = 4.36AGDD16 pKa = 3.69RR17 pKa = 11.84IYY19 pKa = 10.72PGLDD23 pKa = 2.63ARR25 pKa = 11.84QRR27 pKa = 11.84AARR30 pKa = 11.84MEE32 pKa = 4.04RR33 pKa = 11.84EE34 pKa = 4.09SFGVHH39 pKa = 4.26TRR41 pKa = 11.84EE42 pKa = 3.87FAQPASAHH50 pKa = 6.48PEE52 pKa = 3.83PSSEE56 pKa = 3.72ARR58 pKa = 11.84EE59 pKa = 4.1VVLAMLEE66 pKa = 4.21QEE68 pKa = 4.74FDD70 pKa = 3.4PSQWQLPDD78 pKa = 3.45DD79 pKa = 3.85WDD81 pKa = 3.92SFDD84 pKa = 3.33SYY86 pKa = 11.63EE87 pKa = 4.44RR88 pKa = 11.84VLSRR92 pKa = 11.84LNMQSSPGYY101 pKa = 8.77PYY103 pKa = 10.93SSQAPTNGEE112 pKa = 3.77FLGWDD117 pKa = 3.26GVEE120 pKa = 4.06CQNRR124 pKa = 11.84QRR126 pKa = 11.84VMILWRR132 pKa = 11.84DD133 pKa = 3.48VKK135 pKa = 10.82RR136 pKa = 11.84LFEE139 pKa = 4.21EE140 pKa = 4.25EE141 pKa = 4.01QVEE144 pKa = 4.74VILTTFVKK152 pKa = 10.58EE153 pKa = 4.23EE154 pKa = 3.73PHH156 pKa = 5.3TLKK159 pKa = 10.86KK160 pKa = 10.4VGARR164 pKa = 11.84RR165 pKa = 11.84WRR167 pKa = 11.84LIQAAPLHH175 pKa = 5.37VQVWWQLLFCYY186 pKa = 10.55GNDD189 pKa = 2.96AMVRR193 pKa = 11.84EE194 pKa = 4.99SINIPAQQGFTIVGGAWKK212 pKa = 10.2SYY214 pKa = 10.15LDD216 pKa = 3.31SWRR219 pKa = 11.84NKK221 pKa = 9.04GLRR224 pKa = 11.84CGLDD228 pKa = 3.75FSAFDD233 pKa = 3.23WTVRR237 pKa = 11.84KK238 pKa = 9.2WMLDD242 pKa = 3.08DD243 pKa = 4.4ALRR246 pKa = 11.84LRR248 pKa = 11.84LALGRR253 pKa = 11.84GRR255 pKa = 11.84RR256 pKa = 11.84LGDD259 pKa = 2.82WHH261 pKa = 6.0RR262 pKa = 11.84QARR265 pKa = 11.84KK266 pKa = 9.31AYY268 pKa = 8.4HH269 pKa = 6.24AMYY272 pKa = 10.53VNPKK276 pKa = 9.84VLFSDD281 pKa = 4.46GSLWEE286 pKa = 3.88QTVPGIQKK294 pKa = 10.0SGVVNTIADD303 pKa = 3.49NSVIGRR309 pKa = 11.84MNYY312 pKa = 8.38LHH314 pKa = 7.11AWLVYY319 pKa = 9.88LAEE322 pKa = 4.82AEE324 pKa = 4.27QVDD327 pKa = 3.88PEE329 pKa = 4.72LYY331 pKa = 9.54QRR333 pKa = 11.84PASVHH338 pKa = 6.53CGDD341 pKa = 5.02DD342 pKa = 3.24MLVASKK348 pKa = 11.12AKK350 pKa = 10.1LLKK353 pKa = 10.17PYY355 pKa = 10.41LSRR358 pKa = 11.84FGLVVKK364 pKa = 10.51YY365 pKa = 8.01LTQGLEE371 pKa = 4.09FCGHH375 pKa = 6.82KK376 pKa = 10.2FRR378 pKa = 11.84SAGMIPMYY386 pKa = 7.51WTKK389 pKa = 9.99HH390 pKa = 4.32VKK392 pKa = 9.67MMMHH396 pKa = 6.47ATDD399 pKa = 3.53VTSSLDD405 pKa = 3.15AMLRR409 pKa = 11.84MYY411 pKa = 10.08AHH413 pKa = 6.73CTKK416 pKa = 10.36KK417 pKa = 9.35WKK419 pKa = 9.68EE420 pKa = 3.55WAALADD426 pKa = 3.73EE427 pKa = 5.17LGVKK431 pKa = 10.2RR432 pKa = 11.84FSVLLYY438 pKa = 9.68RR439 pKa = 11.84HH440 pKa = 5.98WKK442 pKa = 9.75DD443 pKa = 5.4RR444 pKa = 11.84PADD447 pKa = 3.63WSPAAQTLWW456 pKa = 3.28
MM1 pKa = 7.86RR2 pKa = 11.84SVDD5 pKa = 4.46DD6 pKa = 4.11EE7 pKa = 4.78LLFPSEE13 pKa = 4.36AGDD16 pKa = 3.69RR17 pKa = 11.84IYY19 pKa = 10.72PGLDD23 pKa = 2.63ARR25 pKa = 11.84QRR27 pKa = 11.84AARR30 pKa = 11.84MEE32 pKa = 4.04RR33 pKa = 11.84EE34 pKa = 4.09SFGVHH39 pKa = 4.26TRR41 pKa = 11.84EE42 pKa = 3.87FAQPASAHH50 pKa = 6.48PEE52 pKa = 3.83PSSEE56 pKa = 3.72ARR58 pKa = 11.84EE59 pKa = 4.1VVLAMLEE66 pKa = 4.21QEE68 pKa = 4.74FDD70 pKa = 3.4PSQWQLPDD78 pKa = 3.45DD79 pKa = 3.85WDD81 pKa = 3.92SFDD84 pKa = 3.33SYY86 pKa = 11.63EE87 pKa = 4.44RR88 pKa = 11.84VLSRR92 pKa = 11.84LNMQSSPGYY101 pKa = 8.77PYY103 pKa = 10.93SSQAPTNGEE112 pKa = 3.77FLGWDD117 pKa = 3.26GVEE120 pKa = 4.06CQNRR124 pKa = 11.84QRR126 pKa = 11.84VMILWRR132 pKa = 11.84DD133 pKa = 3.48VKK135 pKa = 10.82RR136 pKa = 11.84LFEE139 pKa = 4.21EE140 pKa = 4.25EE141 pKa = 4.01QVEE144 pKa = 4.74VILTTFVKK152 pKa = 10.58EE153 pKa = 4.23EE154 pKa = 3.73PHH156 pKa = 5.3TLKK159 pKa = 10.86KK160 pKa = 10.4VGARR164 pKa = 11.84RR165 pKa = 11.84WRR167 pKa = 11.84LIQAAPLHH175 pKa = 5.37VQVWWQLLFCYY186 pKa = 10.55GNDD189 pKa = 2.96AMVRR193 pKa = 11.84EE194 pKa = 4.99SINIPAQQGFTIVGGAWKK212 pKa = 10.2SYY214 pKa = 10.15LDD216 pKa = 3.31SWRR219 pKa = 11.84NKK221 pKa = 9.04GLRR224 pKa = 11.84CGLDD228 pKa = 3.75FSAFDD233 pKa = 3.23WTVRR237 pKa = 11.84KK238 pKa = 9.2WMLDD242 pKa = 3.08DD243 pKa = 4.4ALRR246 pKa = 11.84LRR248 pKa = 11.84LALGRR253 pKa = 11.84GRR255 pKa = 11.84RR256 pKa = 11.84LGDD259 pKa = 2.82WHH261 pKa = 6.0RR262 pKa = 11.84QARR265 pKa = 11.84KK266 pKa = 9.31AYY268 pKa = 8.4HH269 pKa = 6.24AMYY272 pKa = 10.53VNPKK276 pKa = 9.84VLFSDD281 pKa = 4.46GSLWEE286 pKa = 3.88QTVPGIQKK294 pKa = 10.0SGVVNTIADD303 pKa = 3.49NSVIGRR309 pKa = 11.84MNYY312 pKa = 8.38LHH314 pKa = 7.11AWLVYY319 pKa = 9.88LAEE322 pKa = 4.82AEE324 pKa = 4.27QVDD327 pKa = 3.88PEE329 pKa = 4.72LYY331 pKa = 9.54QRR333 pKa = 11.84PASVHH338 pKa = 6.53CGDD341 pKa = 5.02DD342 pKa = 3.24MLVASKK348 pKa = 11.12AKK350 pKa = 10.1LLKK353 pKa = 10.17PYY355 pKa = 10.41LSRR358 pKa = 11.84FGLVVKK364 pKa = 10.51YY365 pKa = 8.01LTQGLEE371 pKa = 4.09FCGHH375 pKa = 6.82KK376 pKa = 10.2FRR378 pKa = 11.84SAGMIPMYY386 pKa = 7.51WTKK389 pKa = 9.99HH390 pKa = 4.32VKK392 pKa = 9.67MMMHH396 pKa = 6.47ATDD399 pKa = 3.53VTSSLDD405 pKa = 3.15AMLRR409 pKa = 11.84MYY411 pKa = 10.08AHH413 pKa = 6.73CTKK416 pKa = 10.36KK417 pKa = 9.35WKK419 pKa = 9.68EE420 pKa = 3.55WAALADD426 pKa = 3.73EE427 pKa = 5.17LGVKK431 pKa = 10.2RR432 pKa = 11.84FSVLLYY438 pKa = 9.68RR439 pKa = 11.84HH440 pKa = 5.98WKK442 pKa = 9.75DD443 pKa = 5.4RR444 pKa = 11.84PADD447 pKa = 3.63WSPAAQTLWW456 pKa = 3.28
Molecular weight: 52.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
982 |
456 |
526 |
491.0 |
54.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.776 ± 0.816 | 1.935 ± 0.413 |
4.786 ± 0.903 | 6.11 ± 0.126 |
2.953 ± 0.517 | 7.943 ± 1.348 |
2.953 ± 0.068 | 3.259 ± 0.564 |
4.481 ± 0.229 | 9.369 ± 0.48 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.953 ± 0.517 | 1.629 ± 0.376 |
5.397 ± 0.528 | 4.481 ± 0.229 |
6.415 ± 0.987 | 7.23 ± 0.434 |
3.87 ± 0.387 | 8.656 ± 0.946 |
2.648 ± 1.159 | 3.157 ± 0.235 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
