
Capybara microvirus Cap1_SP_59
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.79
Get precalculated fractions of proteins
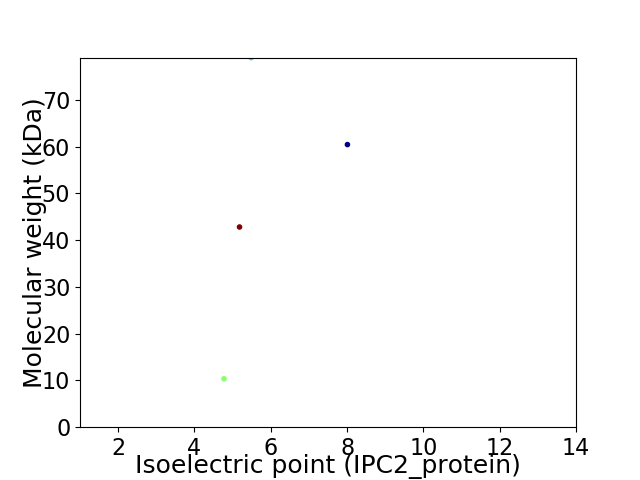
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
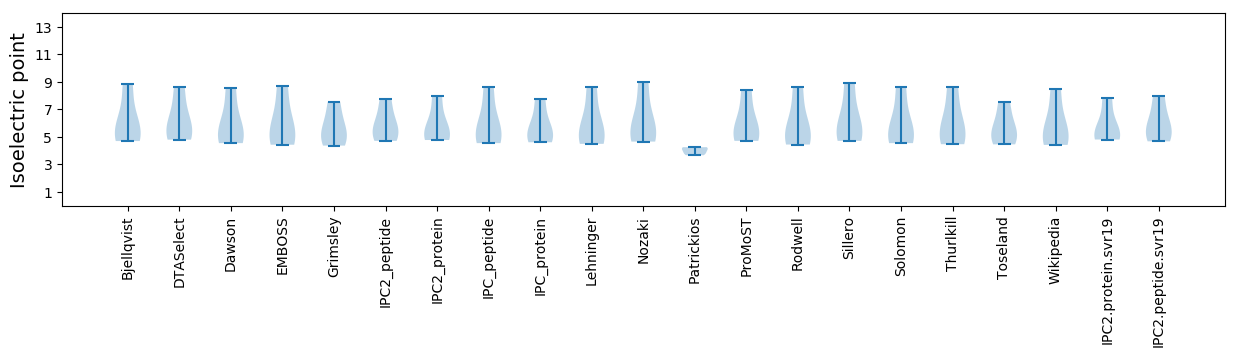
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W3W8|A0A4P8W3W8_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_59 OX=2584784 PE=4 SV=1
MM1 pKa = 6.83NTNLRR6 pKa = 11.84KK7 pKa = 9.36IRR9 pKa = 11.84CNFNPIVEE17 pKa = 4.11VRR19 pKa = 11.84PVKK22 pKa = 10.32IGEE25 pKa = 4.81DD26 pKa = 2.88IDD28 pKa = 3.6IRR30 pKa = 11.84FVDD33 pKa = 5.34PSASDD38 pKa = 3.85LPSPDD43 pKa = 2.94SMRR46 pKa = 11.84LEE48 pKa = 4.31RR49 pKa = 11.84LLSAGYY55 pKa = 10.03RR56 pKa = 11.84PEE58 pKa = 4.09QVSPAFMTPTAAHH71 pKa = 6.04TLEE74 pKa = 5.17DD75 pKa = 3.77MAEE78 pKa = 4.03SAINSIVEE86 pKa = 3.87TSNNQEE92 pKa = 3.91SS93 pKa = 3.55
MM1 pKa = 6.83NTNLRR6 pKa = 11.84KK7 pKa = 9.36IRR9 pKa = 11.84CNFNPIVEE17 pKa = 4.11VRR19 pKa = 11.84PVKK22 pKa = 10.32IGEE25 pKa = 4.81DD26 pKa = 2.88IDD28 pKa = 3.6IRR30 pKa = 11.84FVDD33 pKa = 5.34PSASDD38 pKa = 3.85LPSPDD43 pKa = 2.94SMRR46 pKa = 11.84LEE48 pKa = 4.31RR49 pKa = 11.84LLSAGYY55 pKa = 10.03RR56 pKa = 11.84PEE58 pKa = 4.09QVSPAFMTPTAAHH71 pKa = 6.04TLEE74 pKa = 5.17DD75 pKa = 3.77MAEE78 pKa = 4.03SAINSIVEE86 pKa = 3.87TSNNQEE92 pKa = 3.91SS93 pKa = 3.55
Molecular weight: 10.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W3S1|A0A4P8W3S1_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_59 OX=2584784 PE=4 SV=1
MM1 pKa = 7.85CISKK5 pKa = 10.28IGIRR9 pKa = 11.84SKK11 pKa = 11.19DD12 pKa = 3.46LVYY15 pKa = 11.03NQYY18 pKa = 8.95TPAFRR23 pKa = 11.84SVPCGICPQCLNTKK37 pKa = 9.54RR38 pKa = 11.84NNMYY42 pKa = 9.92IRR44 pKa = 11.84SFFEE48 pKa = 3.74WQHH51 pKa = 4.42TLKK54 pKa = 10.92EE55 pKa = 4.28GGFVLFPTYY64 pKa = 10.08TFQDD68 pKa = 2.76SDD70 pKa = 3.87LPYY73 pKa = 10.43FDD75 pKa = 4.77MRR77 pKa = 11.84DD78 pKa = 3.49GSLHH82 pKa = 6.88RR83 pKa = 11.84GVQDD87 pKa = 3.46SYY89 pKa = 11.94HH90 pKa = 7.36LDD92 pKa = 4.38DD93 pKa = 5.34YY94 pKa = 11.4ICPCFDD100 pKa = 3.54FSDD103 pKa = 3.29IRR105 pKa = 11.84GTKK108 pKa = 10.26GIVKK112 pKa = 10.25AIRR115 pKa = 11.84SHH117 pKa = 7.14ARR119 pKa = 11.84DD120 pKa = 3.1VWNVVGVKK128 pKa = 9.88YY129 pKa = 8.74VVCPEE134 pKa = 4.03YY135 pKa = 11.12GPTPNKK141 pKa = 6.35THH143 pKa = 7.06RR144 pKa = 11.84PHH146 pKa = 6.15FHH148 pKa = 7.15CLFFCPPSPLGSLDD162 pKa = 3.68GDD164 pKa = 3.53LQRR167 pKa = 11.84RR168 pKa = 11.84YY169 pKa = 10.0KK170 pKa = 11.14DD171 pKa = 4.06DD172 pKa = 3.41MAKK175 pKa = 10.38FCLKK179 pKa = 9.95VWSEE183 pKa = 4.24RR184 pKa = 11.84KK185 pKa = 9.18GHH187 pKa = 5.44GTASYY192 pKa = 9.77SKK194 pKa = 10.33KK195 pKa = 9.58YY196 pKa = 10.87GPFIEE201 pKa = 4.33SFKK204 pKa = 9.83GIKK207 pKa = 9.31YY208 pKa = 9.41ASKK211 pKa = 11.05YY212 pKa = 4.71MTKK215 pKa = 10.57DD216 pKa = 3.42VVDD219 pKa = 3.96LCEE222 pKa = 3.95NCPRR226 pKa = 11.84LKK228 pKa = 10.63KK229 pKa = 10.12FLHH232 pKa = 6.52HH233 pKa = 5.91NTYY236 pKa = 10.72DD237 pKa = 3.56NKK239 pKa = 10.28VFLPDD244 pKa = 2.96NWQEE248 pKa = 3.7IPLYY252 pKa = 9.18EE253 pKa = 4.62LPYY256 pKa = 9.21QYY258 pKa = 10.24RR259 pKa = 11.84YY260 pKa = 10.24KK261 pKa = 10.55AIKK264 pKa = 9.74PFLPRR269 pKa = 11.84PLWCKK274 pKa = 10.55GFGEE278 pKa = 5.46SILQLLTDD286 pKa = 3.88STDD289 pKa = 3.41EE290 pKa = 3.96QLTDD294 pKa = 4.13LLVNGIKK301 pKa = 9.47IDD303 pKa = 3.84GDD305 pKa = 3.19ISLDD309 pKa = 3.54GKK311 pKa = 10.75QNTYY315 pKa = 10.49QIPRR319 pKa = 11.84YY320 pKa = 8.8IQQKK324 pKa = 6.86FTWHH328 pKa = 6.23TFDD331 pKa = 5.61GYY333 pKa = 11.06DD334 pKa = 3.1IYY336 pKa = 11.52NSRR339 pKa = 11.84LSRR342 pKa = 11.84ALRR345 pKa = 11.84NVYY348 pKa = 7.16EE349 pKa = 3.78QRR351 pKa = 11.84LNQKK355 pKa = 10.07SLLLRR360 pKa = 11.84NFFYY364 pKa = 10.96NSHH367 pKa = 6.9KK368 pKa = 10.8YY369 pKa = 7.1MHH371 pKa = 6.99IWNDD375 pKa = 3.07LHH377 pKa = 9.12SPFDD381 pKa = 4.19LKK383 pKa = 11.3DD384 pKa = 3.47LLSRR388 pKa = 11.84ISPTQLACYY397 pKa = 9.87SEE399 pKa = 4.26IMCTNFAKK407 pKa = 10.55SDD409 pKa = 3.78FEE411 pKa = 5.88DD412 pKa = 3.83INSLPFPDD420 pKa = 4.01VEE422 pKa = 4.36YY423 pKa = 11.37KK424 pKa = 10.91EE425 pKa = 4.19MTLKK429 pKa = 11.02YY430 pKa = 8.58MLDD433 pKa = 3.53RR434 pKa = 11.84IIYY437 pKa = 7.25HH438 pKa = 6.4KK439 pKa = 10.64PNIKK443 pKa = 10.07CLPHH447 pKa = 6.62LKK449 pKa = 10.7GLFKK453 pKa = 10.44PLEE456 pKa = 3.78AHH458 pKa = 6.63KK459 pKa = 10.29PYY461 pKa = 10.73INDD464 pKa = 3.36EE465 pKa = 4.2FSLLLHH471 pKa = 6.82FYY473 pKa = 9.39SKK475 pKa = 11.02CMQEE479 pKa = 4.67DD480 pKa = 4.02VNKK483 pKa = 10.13QLYY486 pKa = 9.37LRR488 pKa = 11.84RR489 pKa = 11.84KK490 pKa = 8.74KK491 pKa = 9.95RR492 pKa = 11.84HH493 pKa = 6.07HH494 pKa = 6.68DD495 pKa = 4.16AIVQAEE501 pKa = 4.46LLPTLIKK508 pKa = 10.76YY509 pKa = 9.28EE510 pKa = 4.18YY511 pKa = 10.82
MM1 pKa = 7.85CISKK5 pKa = 10.28IGIRR9 pKa = 11.84SKK11 pKa = 11.19DD12 pKa = 3.46LVYY15 pKa = 11.03NQYY18 pKa = 8.95TPAFRR23 pKa = 11.84SVPCGICPQCLNTKK37 pKa = 9.54RR38 pKa = 11.84NNMYY42 pKa = 9.92IRR44 pKa = 11.84SFFEE48 pKa = 3.74WQHH51 pKa = 4.42TLKK54 pKa = 10.92EE55 pKa = 4.28GGFVLFPTYY64 pKa = 10.08TFQDD68 pKa = 2.76SDD70 pKa = 3.87LPYY73 pKa = 10.43FDD75 pKa = 4.77MRR77 pKa = 11.84DD78 pKa = 3.49GSLHH82 pKa = 6.88RR83 pKa = 11.84GVQDD87 pKa = 3.46SYY89 pKa = 11.94HH90 pKa = 7.36LDD92 pKa = 4.38DD93 pKa = 5.34YY94 pKa = 11.4ICPCFDD100 pKa = 3.54FSDD103 pKa = 3.29IRR105 pKa = 11.84GTKK108 pKa = 10.26GIVKK112 pKa = 10.25AIRR115 pKa = 11.84SHH117 pKa = 7.14ARR119 pKa = 11.84DD120 pKa = 3.1VWNVVGVKK128 pKa = 9.88YY129 pKa = 8.74VVCPEE134 pKa = 4.03YY135 pKa = 11.12GPTPNKK141 pKa = 6.35THH143 pKa = 7.06RR144 pKa = 11.84PHH146 pKa = 6.15FHH148 pKa = 7.15CLFFCPPSPLGSLDD162 pKa = 3.68GDD164 pKa = 3.53LQRR167 pKa = 11.84RR168 pKa = 11.84YY169 pKa = 10.0KK170 pKa = 11.14DD171 pKa = 4.06DD172 pKa = 3.41MAKK175 pKa = 10.38FCLKK179 pKa = 9.95VWSEE183 pKa = 4.24RR184 pKa = 11.84KK185 pKa = 9.18GHH187 pKa = 5.44GTASYY192 pKa = 9.77SKK194 pKa = 10.33KK195 pKa = 9.58YY196 pKa = 10.87GPFIEE201 pKa = 4.33SFKK204 pKa = 9.83GIKK207 pKa = 9.31YY208 pKa = 9.41ASKK211 pKa = 11.05YY212 pKa = 4.71MTKK215 pKa = 10.57DD216 pKa = 3.42VVDD219 pKa = 3.96LCEE222 pKa = 3.95NCPRR226 pKa = 11.84LKK228 pKa = 10.63KK229 pKa = 10.12FLHH232 pKa = 6.52HH233 pKa = 5.91NTYY236 pKa = 10.72DD237 pKa = 3.56NKK239 pKa = 10.28VFLPDD244 pKa = 2.96NWQEE248 pKa = 3.7IPLYY252 pKa = 9.18EE253 pKa = 4.62LPYY256 pKa = 9.21QYY258 pKa = 10.24RR259 pKa = 11.84YY260 pKa = 10.24KK261 pKa = 10.55AIKK264 pKa = 9.74PFLPRR269 pKa = 11.84PLWCKK274 pKa = 10.55GFGEE278 pKa = 5.46SILQLLTDD286 pKa = 3.88STDD289 pKa = 3.41EE290 pKa = 3.96QLTDD294 pKa = 4.13LLVNGIKK301 pKa = 9.47IDD303 pKa = 3.84GDD305 pKa = 3.19ISLDD309 pKa = 3.54GKK311 pKa = 10.75QNTYY315 pKa = 10.49QIPRR319 pKa = 11.84YY320 pKa = 8.8IQQKK324 pKa = 6.86FTWHH328 pKa = 6.23TFDD331 pKa = 5.61GYY333 pKa = 11.06DD334 pKa = 3.1IYY336 pKa = 11.52NSRR339 pKa = 11.84LSRR342 pKa = 11.84ALRR345 pKa = 11.84NVYY348 pKa = 7.16EE349 pKa = 3.78QRR351 pKa = 11.84LNQKK355 pKa = 10.07SLLLRR360 pKa = 11.84NFFYY364 pKa = 10.96NSHH367 pKa = 6.9KK368 pKa = 10.8YY369 pKa = 7.1MHH371 pKa = 6.99IWNDD375 pKa = 3.07LHH377 pKa = 9.12SPFDD381 pKa = 4.19LKK383 pKa = 11.3DD384 pKa = 3.47LLSRR388 pKa = 11.84ISPTQLACYY397 pKa = 9.87SEE399 pKa = 4.26IMCTNFAKK407 pKa = 10.55SDD409 pKa = 3.78FEE411 pKa = 5.88DD412 pKa = 3.83INSLPFPDD420 pKa = 4.01VEE422 pKa = 4.36YY423 pKa = 11.37KK424 pKa = 10.91EE425 pKa = 4.19MTLKK429 pKa = 11.02YY430 pKa = 8.58MLDD433 pKa = 3.53RR434 pKa = 11.84IIYY437 pKa = 7.25HH438 pKa = 6.4KK439 pKa = 10.64PNIKK443 pKa = 10.07CLPHH447 pKa = 6.62LKK449 pKa = 10.7GLFKK453 pKa = 10.44PLEE456 pKa = 3.78AHH458 pKa = 6.63KK459 pKa = 10.29PYY461 pKa = 10.73INDD464 pKa = 3.36EE465 pKa = 4.2FSLLLHH471 pKa = 6.82FYY473 pKa = 9.39SKK475 pKa = 11.02CMQEE479 pKa = 4.67DD480 pKa = 4.02VNKK483 pKa = 10.13QLYY486 pKa = 9.37LRR488 pKa = 11.84RR489 pKa = 11.84KK490 pKa = 8.74KK491 pKa = 9.95RR492 pKa = 11.84HH493 pKa = 6.07HH494 pKa = 6.68DD495 pKa = 4.16AIVQAEE501 pKa = 4.46LLPTLIKK508 pKa = 10.76YY509 pKa = 9.28EE510 pKa = 4.18YY511 pKa = 10.82
Molecular weight: 60.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1687 |
93 |
695 |
421.8 |
48.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.343 ± 2.583 | 1.6 ± 0.624 |
6.461 ± 0.836 | 5.216 ± 1.096 |
5.039 ± 1.04 | 4.861 ± 0.233 |
2.134 ± 0.576 | 5.098 ± 0.369 |
6.52 ± 0.671 | 9.01 ± 0.634 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.253 ± 0.369 | 6.165 ± 0.873 |
4.149 ± 0.866 | 5.157 ± 1.406 |
4.505 ± 0.413 | 8.536 ± 1.107 |
5.75 ± 0.983 | 4.683 ± 0.88 |
0.948 ± 0.147 | 5.572 ± 0.818 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
