
Lloviu cuevavirus (isolate Bat/Spain/Asturias-Bat86/2003)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Filoviridae; Cuevavirus; Lloviu cuevavirus
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins
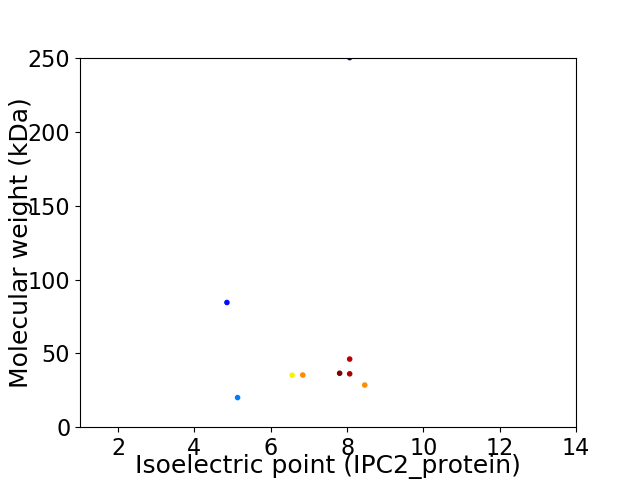
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
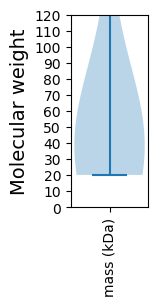
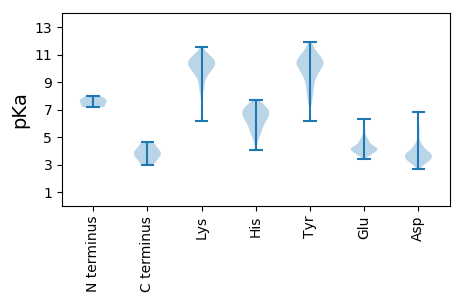
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G8EFI2|G8EFI2_LLOVA Polymerase cofactor VP35 OS=Lloviu cuevavirus (isolate Bat/Spain/Asturias-Bat86/2003) OX=1559366 PE=3 SV=1
MM1 pKa = 7.21NRR3 pKa = 11.84YY4 pKa = 8.91LGHH7 pKa = 5.53GTRR10 pKa = 11.84TSRR13 pKa = 11.84EE14 pKa = 3.84NTNLSEE20 pKa = 3.96LHH22 pKa = 6.72GILSLGLNVDD32 pKa = 3.34HH33 pKa = 7.02TIVRR37 pKa = 11.84KK38 pKa = 10.07KK39 pKa = 10.54SIPLFEE45 pKa = 6.29IGNSDD50 pKa = 4.38QVCNWIIQIIEE61 pKa = 4.34AGVDD65 pKa = 3.71LQDD68 pKa = 3.68VADD71 pKa = 4.29SFLTMLCVNHH81 pKa = 7.52AYY83 pKa = 10.37QGDD86 pKa = 3.94PNLFLEE92 pKa = 5.02SPAAHH97 pKa = 6.21YY98 pKa = 10.69LKK100 pKa = 10.76GHH102 pKa = 7.16GIHH105 pKa = 6.76FEE107 pKa = 3.74IQHH110 pKa = 6.64RR111 pKa = 11.84DD112 pKa = 3.64NVDD115 pKa = 3.73HH116 pKa = 6.74ITDD119 pKa = 4.08LLGVGSRR126 pKa = 11.84DD127 pKa = 2.95KK128 pKa = 11.13SLRR131 pKa = 11.84KK132 pKa = 7.86TLSALEE138 pKa = 4.21FEE140 pKa = 5.27PDD142 pKa = 3.2GSTTAGMFLSFASLFLPKK160 pKa = 10.36LVVGEE165 pKa = 4.29RR166 pKa = 11.84ACLEE170 pKa = 3.85KK171 pKa = 10.79VQRR174 pKa = 11.84QIQIHH179 pKa = 6.28AEE181 pKa = 3.79QGLIQYY187 pKa = 7.39PTQWQSVGHH196 pKa = 5.02MMVVFRR202 pKa = 11.84LIRR205 pKa = 11.84VNFVLKK211 pKa = 10.4FLLVHH216 pKa = 6.24QGMHH220 pKa = 5.31MMAGHH225 pKa = 7.48DD226 pKa = 3.95ANDD229 pKa = 4.4AIIANSISQTRR240 pKa = 11.84FSGLLIVKK248 pKa = 8.18TVLEE252 pKa = 5.03HH253 pKa = 7.17ILQKK257 pKa = 10.01TEE259 pKa = 4.38AGVQLHH265 pKa = 6.13PLARR269 pKa = 11.84TSKK272 pKa = 10.79VKK274 pKa = 11.11GEE276 pKa = 3.92LLAFKK281 pKa = 10.58SALEE285 pKa = 3.83ALASHH290 pKa = 7.08RR291 pKa = 11.84EE292 pKa = 3.95YY293 pKa = 11.47APFARR298 pKa = 11.84LLNLSGVNNLEE309 pKa = 4.28HH310 pKa = 6.95GLYY313 pKa = 9.39PQLSAIALGVATAHH327 pKa = 6.58GSTLAGVNVSEE338 pKa = 5.05QYY340 pKa = 9.67QQLRR344 pKa = 11.84EE345 pKa = 4.06AATEE349 pKa = 4.1AEE351 pKa = 4.48KK352 pKa = 10.71QLQQHH357 pKa = 5.3SEE359 pKa = 3.85MRR361 pKa = 11.84EE362 pKa = 4.03LEE364 pKa = 4.17TLGLDD369 pKa = 3.34EE370 pKa = 4.89QEE372 pKa = 4.29RR373 pKa = 11.84KK374 pKa = 9.7ILATFHH380 pKa = 6.2SRR382 pKa = 11.84KK383 pKa = 9.96NEE385 pKa = 3.52INIQQTSSILAIRR398 pKa = 11.84KK399 pKa = 7.82EE400 pKa = 3.98RR401 pKa = 11.84LRR403 pKa = 11.84KK404 pKa = 8.52LTEE407 pKa = 3.95ALNEE411 pKa = 4.21EE412 pKa = 4.49KK413 pKa = 10.82NKK415 pKa = 10.42NALDD419 pKa = 4.45DD420 pKa = 4.21EE421 pKa = 5.08DD422 pKa = 5.18EE423 pKa = 4.68SEE425 pKa = 4.24EE426 pKa = 5.15DD427 pKa = 3.42DD428 pKa = 3.44WSPEE432 pKa = 3.61NRR434 pKa = 11.84GIRR437 pKa = 11.84SNKK440 pKa = 9.56GSTKK444 pKa = 10.16EE445 pKa = 4.03SSSYY449 pKa = 8.5TASRR453 pKa = 11.84TEE455 pKa = 3.8EE456 pKa = 3.72DD457 pKa = 3.46RR458 pKa = 11.84NNYY461 pKa = 8.45NSKK464 pKa = 10.57DD465 pKa = 3.42DD466 pKa = 3.96HH467 pKa = 7.59LSGKK471 pKa = 8.05EE472 pKa = 3.87QMSTQQEE479 pKa = 4.33SGADD483 pKa = 3.7DD484 pKa = 5.59LDD486 pKa = 5.68LFDD489 pKa = 6.82LDD491 pKa = 6.14DD492 pKa = 6.19DD493 pKa = 5.49GDD495 pKa = 4.32TNSQDD500 pKa = 3.2PNNRR504 pKa = 11.84QKK506 pKa = 10.81QSDD509 pKa = 3.86TQQTQEE515 pKa = 3.72SSDD518 pKa = 3.36RR519 pKa = 11.84SDD521 pKa = 3.4YY522 pKa = 11.07SRR524 pKa = 11.84RR525 pKa = 11.84PAYY528 pKa = 9.7DD529 pKa = 3.28WPPGDD534 pKa = 4.84RR535 pKa = 11.84PHH537 pKa = 6.25TTQATDD543 pKa = 2.78EE544 pKa = 4.44HH545 pKa = 6.69TDD547 pKa = 3.96LLNKK551 pKa = 9.32DD552 pKa = 3.24HH553 pKa = 7.36RR554 pKa = 11.84RR555 pKa = 11.84NQVKK559 pKa = 9.86PGRR562 pKa = 11.84RR563 pKa = 11.84GNDD566 pKa = 3.0PRR568 pKa = 11.84TLPLISFDD576 pKa = 4.27DD577 pKa = 4.1NEE579 pKa = 5.12GEE581 pKa = 4.22ILDD584 pKa = 4.99DD585 pKa = 5.54KK586 pKa = 11.54SDD588 pKa = 3.8LPAPDD593 pKa = 3.54THH595 pKa = 7.21SDD597 pKa = 3.43PDD599 pKa = 3.84TEE601 pKa = 4.33EE602 pKa = 4.64SEE604 pKa = 4.71EE605 pKa = 3.99EE606 pKa = 4.28HH607 pKa = 6.93PDD609 pKa = 3.86EE610 pKa = 4.85EE611 pKa = 4.91LLPPAPKK618 pKa = 10.77YY619 pKa = 7.79NTKK622 pKa = 9.98TSEE625 pKa = 3.99QEE627 pKa = 3.73PGDD630 pKa = 3.69WKK632 pKa = 11.1QPTSPLSTIPEE643 pKa = 4.09EE644 pKa = 4.46EE645 pKa = 4.69GGHH648 pKa = 5.56EE649 pKa = 4.06ANNDD653 pKa = 3.25NSEE656 pKa = 4.1SDD658 pKa = 5.62LIDD661 pKa = 3.55QMYY664 pKa = 9.59QHH666 pKa = 6.88IFEE669 pKa = 4.7TEE671 pKa = 3.46GAYY674 pKa = 10.18AAINYY679 pKa = 7.99YY680 pKa = 10.61YY681 pKa = 9.06KK682 pKa = 8.5TTGRR686 pKa = 11.84PVTFTSNNNHH696 pKa = 7.38DD697 pKa = 3.72YY698 pKa = 10.63TFPQDD703 pKa = 3.33IEE705 pKa = 4.75GLFPPWEE712 pKa = 4.21GKK714 pKa = 9.85EE715 pKa = 3.98NQKK718 pKa = 10.1VAEE721 pKa = 4.13ILTNSLHH728 pKa = 5.62EE729 pKa = 4.7TGQEE733 pKa = 3.82WADD736 pKa = 3.43MSAKK740 pKa = 9.54EE741 pKa = 4.54RR742 pKa = 11.84YY743 pKa = 9.52LFLINNN749 pKa = 4.44
MM1 pKa = 7.21NRR3 pKa = 11.84YY4 pKa = 8.91LGHH7 pKa = 5.53GTRR10 pKa = 11.84TSRR13 pKa = 11.84EE14 pKa = 3.84NTNLSEE20 pKa = 3.96LHH22 pKa = 6.72GILSLGLNVDD32 pKa = 3.34HH33 pKa = 7.02TIVRR37 pKa = 11.84KK38 pKa = 10.07KK39 pKa = 10.54SIPLFEE45 pKa = 6.29IGNSDD50 pKa = 4.38QVCNWIIQIIEE61 pKa = 4.34AGVDD65 pKa = 3.71LQDD68 pKa = 3.68VADD71 pKa = 4.29SFLTMLCVNHH81 pKa = 7.52AYY83 pKa = 10.37QGDD86 pKa = 3.94PNLFLEE92 pKa = 5.02SPAAHH97 pKa = 6.21YY98 pKa = 10.69LKK100 pKa = 10.76GHH102 pKa = 7.16GIHH105 pKa = 6.76FEE107 pKa = 3.74IQHH110 pKa = 6.64RR111 pKa = 11.84DD112 pKa = 3.64NVDD115 pKa = 3.73HH116 pKa = 6.74ITDD119 pKa = 4.08LLGVGSRR126 pKa = 11.84DD127 pKa = 2.95KK128 pKa = 11.13SLRR131 pKa = 11.84KK132 pKa = 7.86TLSALEE138 pKa = 4.21FEE140 pKa = 5.27PDD142 pKa = 3.2GSTTAGMFLSFASLFLPKK160 pKa = 10.36LVVGEE165 pKa = 4.29RR166 pKa = 11.84ACLEE170 pKa = 3.85KK171 pKa = 10.79VQRR174 pKa = 11.84QIQIHH179 pKa = 6.28AEE181 pKa = 3.79QGLIQYY187 pKa = 7.39PTQWQSVGHH196 pKa = 5.02MMVVFRR202 pKa = 11.84LIRR205 pKa = 11.84VNFVLKK211 pKa = 10.4FLLVHH216 pKa = 6.24QGMHH220 pKa = 5.31MMAGHH225 pKa = 7.48DD226 pKa = 3.95ANDD229 pKa = 4.4AIIANSISQTRR240 pKa = 11.84FSGLLIVKK248 pKa = 8.18TVLEE252 pKa = 5.03HH253 pKa = 7.17ILQKK257 pKa = 10.01TEE259 pKa = 4.38AGVQLHH265 pKa = 6.13PLARR269 pKa = 11.84TSKK272 pKa = 10.79VKK274 pKa = 11.11GEE276 pKa = 3.92LLAFKK281 pKa = 10.58SALEE285 pKa = 3.83ALASHH290 pKa = 7.08RR291 pKa = 11.84EE292 pKa = 3.95YY293 pKa = 11.47APFARR298 pKa = 11.84LLNLSGVNNLEE309 pKa = 4.28HH310 pKa = 6.95GLYY313 pKa = 9.39PQLSAIALGVATAHH327 pKa = 6.58GSTLAGVNVSEE338 pKa = 5.05QYY340 pKa = 9.67QQLRR344 pKa = 11.84EE345 pKa = 4.06AATEE349 pKa = 4.1AEE351 pKa = 4.48KK352 pKa = 10.71QLQQHH357 pKa = 5.3SEE359 pKa = 3.85MRR361 pKa = 11.84EE362 pKa = 4.03LEE364 pKa = 4.17TLGLDD369 pKa = 3.34EE370 pKa = 4.89QEE372 pKa = 4.29RR373 pKa = 11.84KK374 pKa = 9.7ILATFHH380 pKa = 6.2SRR382 pKa = 11.84KK383 pKa = 9.96NEE385 pKa = 3.52INIQQTSSILAIRR398 pKa = 11.84KK399 pKa = 7.82EE400 pKa = 3.98RR401 pKa = 11.84LRR403 pKa = 11.84KK404 pKa = 8.52LTEE407 pKa = 3.95ALNEE411 pKa = 4.21EE412 pKa = 4.49KK413 pKa = 10.82NKK415 pKa = 10.42NALDD419 pKa = 4.45DD420 pKa = 4.21EE421 pKa = 5.08DD422 pKa = 5.18EE423 pKa = 4.68SEE425 pKa = 4.24EE426 pKa = 5.15DD427 pKa = 3.42DD428 pKa = 3.44WSPEE432 pKa = 3.61NRR434 pKa = 11.84GIRR437 pKa = 11.84SNKK440 pKa = 9.56GSTKK444 pKa = 10.16EE445 pKa = 4.03SSSYY449 pKa = 8.5TASRR453 pKa = 11.84TEE455 pKa = 3.8EE456 pKa = 3.72DD457 pKa = 3.46RR458 pKa = 11.84NNYY461 pKa = 8.45NSKK464 pKa = 10.57DD465 pKa = 3.42DD466 pKa = 3.96HH467 pKa = 7.59LSGKK471 pKa = 8.05EE472 pKa = 3.87QMSTQQEE479 pKa = 4.33SGADD483 pKa = 3.7DD484 pKa = 5.59LDD486 pKa = 5.68LFDD489 pKa = 6.82LDD491 pKa = 6.14DD492 pKa = 6.19DD493 pKa = 5.49GDD495 pKa = 4.32TNSQDD500 pKa = 3.2PNNRR504 pKa = 11.84QKK506 pKa = 10.81QSDD509 pKa = 3.86TQQTQEE515 pKa = 3.72SSDD518 pKa = 3.36RR519 pKa = 11.84SDD521 pKa = 3.4YY522 pKa = 11.07SRR524 pKa = 11.84RR525 pKa = 11.84PAYY528 pKa = 9.7DD529 pKa = 3.28WPPGDD534 pKa = 4.84RR535 pKa = 11.84PHH537 pKa = 6.25TTQATDD543 pKa = 2.78EE544 pKa = 4.44HH545 pKa = 6.69TDD547 pKa = 3.96LLNKK551 pKa = 9.32DD552 pKa = 3.24HH553 pKa = 7.36RR554 pKa = 11.84RR555 pKa = 11.84NQVKK559 pKa = 9.86PGRR562 pKa = 11.84RR563 pKa = 11.84GNDD566 pKa = 3.0PRR568 pKa = 11.84TLPLISFDD576 pKa = 4.27DD577 pKa = 4.1NEE579 pKa = 5.12GEE581 pKa = 4.22ILDD584 pKa = 4.99DD585 pKa = 5.54KK586 pKa = 11.54SDD588 pKa = 3.8LPAPDD593 pKa = 3.54THH595 pKa = 7.21SDD597 pKa = 3.43PDD599 pKa = 3.84TEE601 pKa = 4.33EE602 pKa = 4.64SEE604 pKa = 4.71EE605 pKa = 3.99EE606 pKa = 4.28HH607 pKa = 6.93PDD609 pKa = 3.86EE610 pKa = 4.85EE611 pKa = 4.91LLPPAPKK618 pKa = 10.77YY619 pKa = 7.79NTKK622 pKa = 9.98TSEE625 pKa = 3.99QEE627 pKa = 3.73PGDD630 pKa = 3.69WKK632 pKa = 11.1QPTSPLSTIPEE643 pKa = 4.09EE644 pKa = 4.46EE645 pKa = 4.69GGHH648 pKa = 5.56EE649 pKa = 4.06ANNDD653 pKa = 3.25NSEE656 pKa = 4.1SDD658 pKa = 5.62LIDD661 pKa = 3.55QMYY664 pKa = 9.59QHH666 pKa = 6.88IFEE669 pKa = 4.7TEE671 pKa = 3.46GAYY674 pKa = 10.18AAINYY679 pKa = 7.99YY680 pKa = 10.61YY681 pKa = 9.06KK682 pKa = 8.5TTGRR686 pKa = 11.84PVTFTSNNNHH696 pKa = 7.38DD697 pKa = 3.72YY698 pKa = 10.63TFPQDD703 pKa = 3.33IEE705 pKa = 4.75GLFPPWEE712 pKa = 4.21GKK714 pKa = 9.85EE715 pKa = 3.98NQKK718 pKa = 10.1VAEE721 pKa = 4.13ILTNSLHH728 pKa = 5.62EE729 pKa = 4.7TGQEE733 pKa = 3.82WADD736 pKa = 3.43MSAKK740 pKa = 9.54EE741 pKa = 4.54RR742 pKa = 11.84YY743 pKa = 9.52LFLINNN749 pKa = 4.44
Molecular weight: 84.52 kDa
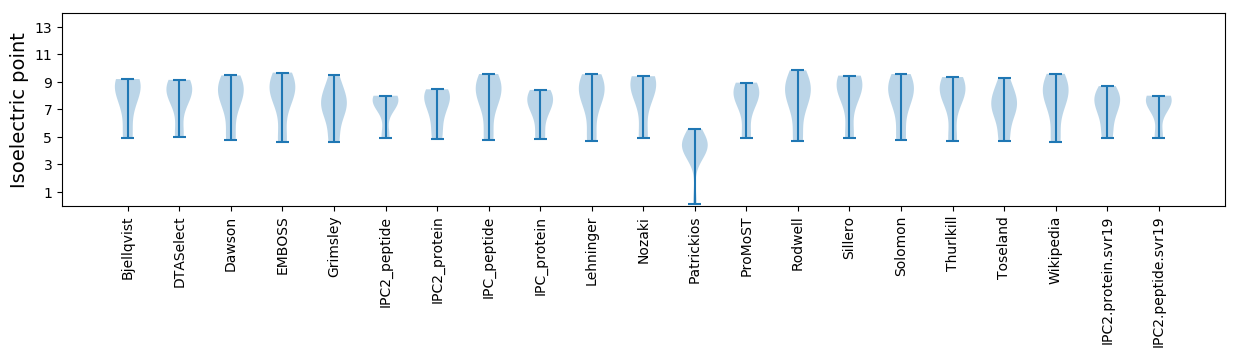
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G8EFI9|G8EFI9_LLOVA RNA-directed RNA polymerase L OS=Lloviu cuevavirus (isolate Bat/Spain/Asturias-Bat86/2003) OX=1559366 PE=3 SV=1
MM1 pKa = 7.51ARR3 pKa = 11.84PSGRR7 pKa = 11.84YY8 pKa = 9.24NILQTDD14 pKa = 4.17TFHH17 pKa = 6.89EE18 pKa = 4.38NTVVKK23 pKa = 10.56HH24 pKa = 5.83DD25 pKa = 3.91LCNFLVTTTITGWDD39 pKa = 4.09VYY41 pKa = 9.68WAGHH45 pKa = 6.71LFHH48 pKa = 6.95VPNKK52 pKa = 10.29GIALLTRR59 pKa = 11.84LKK61 pKa = 10.22TSDD64 pKa = 4.29FAPAWTLTKK73 pKa = 10.82NLFPHH78 pKa = 7.2LFHH81 pKa = 7.27NPTSTLKK88 pKa = 11.08DD89 pKa = 3.74PVWGLRR95 pKa = 11.84VILAAALYY103 pKa = 9.02DD104 pKa = 3.62QVMEE108 pKa = 4.07QSLIKK113 pKa = 10.0PLSEE117 pKa = 3.86TLTLLGDD124 pKa = 3.64WLLTTEE130 pKa = 4.22TQKK133 pKa = 10.93FNLRR137 pKa = 11.84TPRR140 pKa = 11.84AQEE143 pKa = 3.9QISLRR148 pKa = 11.84MLSLVKK154 pKa = 10.45KK155 pKa = 10.01HH156 pKa = 5.81VDD158 pKa = 3.19NFILKK163 pKa = 10.18LLEE166 pKa = 3.69LHH168 pKa = 6.84VINNRR173 pKa = 11.84GFTSSIEE180 pKa = 3.94IGNKK184 pKa = 9.43CNTIIISRR192 pKa = 11.84TNMGYY197 pKa = 10.27LVEE200 pKa = 4.08QQEE203 pKa = 4.31PDD205 pKa = 3.03KK206 pKa = 11.42SALRR210 pKa = 11.84KK211 pKa = 7.52TKK213 pKa = 9.94PGPVKK218 pKa = 9.85FTLVHH223 pKa = 6.47EE224 pKa = 4.45DD225 pKa = 2.91AFRR228 pKa = 11.84DD229 pKa = 4.68FKK231 pKa = 10.99PEE233 pKa = 3.64RR234 pKa = 11.84SSINLLIMEE243 pKa = 5.2FNSSLAII250 pKa = 4.01
MM1 pKa = 7.51ARR3 pKa = 11.84PSGRR7 pKa = 11.84YY8 pKa = 9.24NILQTDD14 pKa = 4.17TFHH17 pKa = 6.89EE18 pKa = 4.38NTVVKK23 pKa = 10.56HH24 pKa = 5.83DD25 pKa = 3.91LCNFLVTTTITGWDD39 pKa = 4.09VYY41 pKa = 9.68WAGHH45 pKa = 6.71LFHH48 pKa = 6.95VPNKK52 pKa = 10.29GIALLTRR59 pKa = 11.84LKK61 pKa = 10.22TSDD64 pKa = 4.29FAPAWTLTKK73 pKa = 10.82NLFPHH78 pKa = 7.2LFHH81 pKa = 7.27NPTSTLKK88 pKa = 11.08DD89 pKa = 3.74PVWGLRR95 pKa = 11.84VILAAALYY103 pKa = 9.02DD104 pKa = 3.62QVMEE108 pKa = 4.07QSLIKK113 pKa = 10.0PLSEE117 pKa = 3.86TLTLLGDD124 pKa = 3.64WLLTTEE130 pKa = 4.22TQKK133 pKa = 10.93FNLRR137 pKa = 11.84TPRR140 pKa = 11.84AQEE143 pKa = 3.9QISLRR148 pKa = 11.84MLSLVKK154 pKa = 10.45KK155 pKa = 10.01HH156 pKa = 5.81VDD158 pKa = 3.19NFILKK163 pKa = 10.18LLEE166 pKa = 3.69LHH168 pKa = 6.84VINNRR173 pKa = 11.84GFTSSIEE180 pKa = 3.94IGNKK184 pKa = 9.43CNTIIISRR192 pKa = 11.84TNMGYY197 pKa = 10.27LVEE200 pKa = 4.08QQEE203 pKa = 4.31PDD205 pKa = 3.03KK206 pKa = 11.42SALRR210 pKa = 11.84KK211 pKa = 7.52TKK213 pKa = 9.94PGPVKK218 pKa = 9.85FTLVHH223 pKa = 6.47EE224 pKa = 4.45DD225 pKa = 2.91AFRR228 pKa = 11.84DD229 pKa = 4.68FKK231 pKa = 10.99PEE233 pKa = 3.64RR234 pKa = 11.84SSINLLIMEE243 pKa = 5.2FNSSLAII250 pKa = 4.01
Molecular weight: 28.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5096 |
189 |
2196 |
566.2 |
63.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.221 ± 0.353 | 1.982 ± 0.429 |
4.749 ± 0.502 | 5.475 ± 0.646 |
3.65 ± 0.548 | 5.926 ± 0.832 |
3.159 ± 0.293 | 5.73 ± 0.547 |
4.847 ± 0.471 | 11.676 ± 0.783 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.805 ± 0.096 | 4.553 ± 0.469 |
5.946 ± 0.713 | 4.611 ± 0.473 |
5.514 ± 0.35 | 7.673 ± 0.464 |
6.888 ± 0.804 | 4.827 ± 0.442 |
1.648 ± 0.349 | 3.12 ± 0.559 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
