
Sewage-associated circular DNA virus-36
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
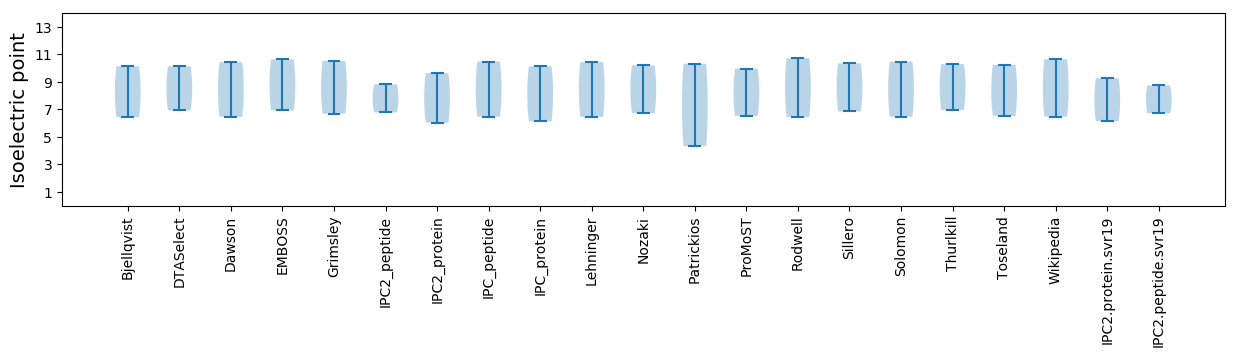
Average proteome isoelectric point is 7.7
Get precalculated fractions of proteins
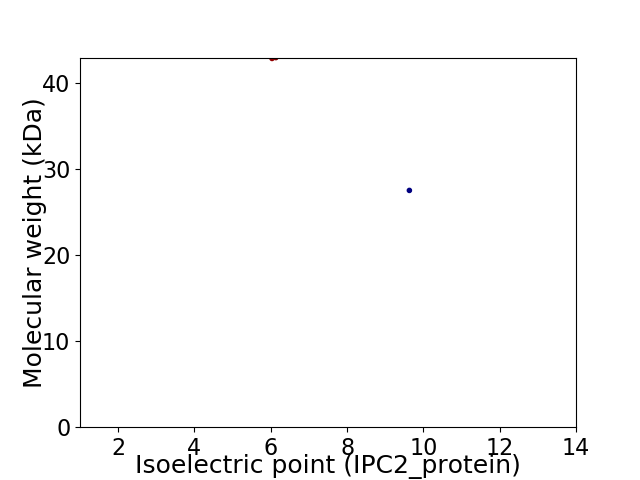
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UH70|A0A0B4UH70_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-36 OX=1592103 PE=3 SV=1
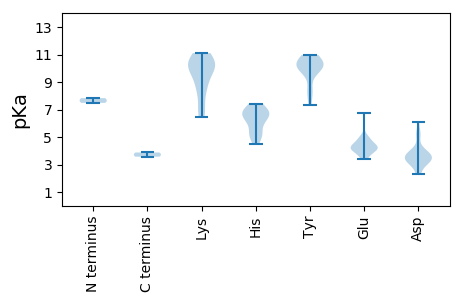
MM1 pKa = 7.48ARR3 pKa = 11.84TPRR6 pKa = 11.84TNAQRR11 pKa = 11.84YY12 pKa = 7.35FLTYY16 pKa = 10.01SQAHH20 pKa = 6.57ALDD23 pKa = 4.21IDD25 pKa = 3.91DD26 pKa = 5.26LANFLHH32 pKa = 6.82ALAPSWLEE40 pKa = 3.5IVQEE44 pKa = 3.95NHH46 pKa = 5.83QDD48 pKa = 3.62DD49 pKa = 5.41GIHH52 pKa = 5.18YY53 pKa = 10.11HH54 pKa = 6.3VVLCFDD60 pKa = 3.73EE61 pKa = 5.0RR62 pKa = 11.84FQRR65 pKa = 11.84ALDD68 pKa = 3.86VFDD71 pKa = 6.06LDD73 pKa = 3.61NHH75 pKa = 6.59HH76 pKa = 7.27PNIAIIKK83 pKa = 8.61NASGDD88 pKa = 3.86LNNRR92 pKa = 11.84RR93 pKa = 11.84HH94 pKa = 6.3YY95 pKa = 9.9IRR97 pKa = 11.84KK98 pKa = 8.85GADD101 pKa = 3.03RR102 pKa = 11.84PKK104 pKa = 10.6EE105 pKa = 4.17SEE107 pKa = 3.96HH108 pKa = 6.77TIADD112 pKa = 3.99HH113 pKa = 7.43KK114 pKa = 10.87KK115 pKa = 9.59GPCDD119 pKa = 3.85YY120 pKa = 10.33IIEE123 pKa = 4.99PDD125 pKa = 3.44TRR127 pKa = 11.84GEE129 pKa = 4.09VPPYY133 pKa = 8.56STSAGRR139 pKa = 11.84LNWGGILEE147 pKa = 4.37SAKK150 pKa = 10.69SEE152 pKa = 4.26EE153 pKa = 4.1EE154 pKa = 3.64FLILVRR160 pKa = 11.84VNQPTEE166 pKa = 3.46WVLRR170 pKa = 11.84NDD172 pKa = 5.18SIVKK176 pKa = 9.1YY177 pKa = 10.96AKK179 pKa = 8.52THH181 pKa = 5.15YY182 pKa = 9.85KK183 pKa = 10.01AARR186 pKa = 11.84EE187 pKa = 4.02PEE189 pKa = 4.12KK190 pKa = 10.9VYY192 pKa = 10.47PPEE195 pKa = 4.0SWVVPPALDD204 pKa = 3.29DD205 pKa = 3.54WVAEE209 pKa = 4.25VFSDD213 pKa = 3.46VSTRR217 pKa = 11.84GVLPRR222 pKa = 11.84PDD224 pKa = 3.45RR225 pKa = 11.84PKK227 pKa = 10.3TLLLVGPTRR236 pKa = 11.84LGKK239 pKa = 9.74SVWAKK244 pKa = 10.08SLGRR248 pKa = 11.84YY249 pKa = 9.67SYY251 pKa = 10.1MCGMWHH257 pKa = 6.6SDD259 pKa = 3.38EE260 pKa = 4.77FDD262 pKa = 3.24EE263 pKa = 4.94SAQYY267 pKa = 10.96LILDD271 pKa = 3.89DD272 pKa = 4.87FEE274 pKa = 6.74FDD276 pKa = 3.69FFHH279 pKa = 7.07GMRR282 pKa = 11.84KK283 pKa = 9.92AIWGAQEE290 pKa = 4.37VFTTTDD296 pKa = 2.3KK297 pKa = 10.13WRR299 pKa = 11.84KK300 pKa = 8.86GVARR304 pKa = 11.84WGKK307 pKa = 6.62PTIWICQEE315 pKa = 3.43EE316 pKa = 4.96HH317 pKa = 6.84NPFTAVNRR325 pKa = 11.84KK326 pKa = 7.7TGLAVMDD333 pKa = 3.47EE334 pKa = 4.61TEE336 pKa = 4.26RR337 pKa = 11.84DD338 pKa = 3.32WYY340 pKa = 10.35KK341 pKa = 10.01QNCVEE346 pKa = 4.07VHH348 pKa = 6.07VNTKK352 pKa = 10.13FIRR355 pKa = 11.84LRR357 pKa = 11.84TCSGRR362 pKa = 11.84SNRR365 pKa = 11.84RR366 pKa = 11.84DD367 pKa = 3.21III369 pKa = 3.88
MM1 pKa = 7.48ARR3 pKa = 11.84TPRR6 pKa = 11.84TNAQRR11 pKa = 11.84YY12 pKa = 7.35FLTYY16 pKa = 10.01SQAHH20 pKa = 6.57ALDD23 pKa = 4.21IDD25 pKa = 3.91DD26 pKa = 5.26LANFLHH32 pKa = 6.82ALAPSWLEE40 pKa = 3.5IVQEE44 pKa = 3.95NHH46 pKa = 5.83QDD48 pKa = 3.62DD49 pKa = 5.41GIHH52 pKa = 5.18YY53 pKa = 10.11HH54 pKa = 6.3VVLCFDD60 pKa = 3.73EE61 pKa = 5.0RR62 pKa = 11.84FQRR65 pKa = 11.84ALDD68 pKa = 3.86VFDD71 pKa = 6.06LDD73 pKa = 3.61NHH75 pKa = 6.59HH76 pKa = 7.27PNIAIIKK83 pKa = 8.61NASGDD88 pKa = 3.86LNNRR92 pKa = 11.84RR93 pKa = 11.84HH94 pKa = 6.3YY95 pKa = 9.9IRR97 pKa = 11.84KK98 pKa = 8.85GADD101 pKa = 3.03RR102 pKa = 11.84PKK104 pKa = 10.6EE105 pKa = 4.17SEE107 pKa = 3.96HH108 pKa = 6.77TIADD112 pKa = 3.99HH113 pKa = 7.43KK114 pKa = 10.87KK115 pKa = 9.59GPCDD119 pKa = 3.85YY120 pKa = 10.33IIEE123 pKa = 4.99PDD125 pKa = 3.44TRR127 pKa = 11.84GEE129 pKa = 4.09VPPYY133 pKa = 8.56STSAGRR139 pKa = 11.84LNWGGILEE147 pKa = 4.37SAKK150 pKa = 10.69SEE152 pKa = 4.26EE153 pKa = 4.1EE154 pKa = 3.64FLILVRR160 pKa = 11.84VNQPTEE166 pKa = 3.46WVLRR170 pKa = 11.84NDD172 pKa = 5.18SIVKK176 pKa = 9.1YY177 pKa = 10.96AKK179 pKa = 8.52THH181 pKa = 5.15YY182 pKa = 9.85KK183 pKa = 10.01AARR186 pKa = 11.84EE187 pKa = 4.02PEE189 pKa = 4.12KK190 pKa = 10.9VYY192 pKa = 10.47PPEE195 pKa = 4.0SWVVPPALDD204 pKa = 3.29DD205 pKa = 3.54WVAEE209 pKa = 4.25VFSDD213 pKa = 3.46VSTRR217 pKa = 11.84GVLPRR222 pKa = 11.84PDD224 pKa = 3.45RR225 pKa = 11.84PKK227 pKa = 10.3TLLLVGPTRR236 pKa = 11.84LGKK239 pKa = 9.74SVWAKK244 pKa = 10.08SLGRR248 pKa = 11.84YY249 pKa = 9.67SYY251 pKa = 10.1MCGMWHH257 pKa = 6.6SDD259 pKa = 3.38EE260 pKa = 4.77FDD262 pKa = 3.24EE263 pKa = 4.94SAQYY267 pKa = 10.96LILDD271 pKa = 3.89DD272 pKa = 4.87FEE274 pKa = 6.74FDD276 pKa = 3.69FFHH279 pKa = 7.07GMRR282 pKa = 11.84KK283 pKa = 9.92AIWGAQEE290 pKa = 4.37VFTTTDD296 pKa = 2.3KK297 pKa = 10.13WRR299 pKa = 11.84KK300 pKa = 8.86GVARR304 pKa = 11.84WGKK307 pKa = 6.62PTIWICQEE315 pKa = 3.43EE316 pKa = 4.96HH317 pKa = 6.84NPFTAVNRR325 pKa = 11.84KK326 pKa = 7.7TGLAVMDD333 pKa = 3.47EE334 pKa = 4.61TEE336 pKa = 4.26RR337 pKa = 11.84DD338 pKa = 3.32WYY340 pKa = 10.35KK341 pKa = 10.01QNCVEE346 pKa = 4.07VHH348 pKa = 6.07VNTKK352 pKa = 10.13FIRR355 pKa = 11.84LRR357 pKa = 11.84TCSGRR362 pKa = 11.84SNRR365 pKa = 11.84RR366 pKa = 11.84DD367 pKa = 3.21III369 pKa = 3.88
Molecular weight: 42.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UH70|A0A0B4UH70_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-36 OX=1592103 PE=3 SV=1
MM1 pKa = 7.83AYY3 pKa = 9.78HH4 pKa = 6.95RR5 pKa = 11.84KK6 pKa = 6.8TRR8 pKa = 11.84RR9 pKa = 11.84GHH11 pKa = 5.3RR12 pKa = 11.84SKK14 pKa = 11.08RR15 pKa = 11.84KK16 pKa = 6.49TPKK19 pKa = 10.05RR20 pKa = 11.84LSQHH24 pKa = 4.48MVRR27 pKa = 11.84AIKK30 pKa = 10.31AISQGEE36 pKa = 4.52VEE38 pKa = 4.71TKK40 pKa = 10.21RR41 pKa = 11.84YY42 pKa = 7.96MVSTDD47 pKa = 2.43ISTALTAAGYY57 pKa = 10.51VSGNNAVIRR66 pKa = 11.84RR67 pKa = 11.84NFLSDD72 pKa = 4.17LPRR75 pKa = 11.84EE76 pKa = 4.33SNTAVNSDD84 pKa = 2.96ASFIGDD90 pKa = 3.55QINLRR95 pKa = 11.84GLRR98 pKa = 11.84WSFHH102 pKa = 5.01GSILPTVNPDD112 pKa = 2.28AMYY115 pKa = 10.83RR116 pKa = 11.84FTVYY120 pKa = 10.27RR121 pKa = 11.84DD122 pKa = 3.14SFYY125 pKa = 10.9FAGTNGPGPGDD136 pKa = 3.79RR137 pKa = 11.84IFEE140 pKa = 4.3QQQSTIPTWSMWNTQVTDD158 pKa = 3.09IVFQRR163 pKa = 11.84KK164 pKa = 7.09FRR166 pKa = 11.84MRR168 pKa = 11.84VSSIGSEE175 pKa = 4.05VLDD178 pKa = 3.49KK179 pKa = 11.1KK180 pKa = 10.85FYY182 pKa = 10.18IPLRR186 pKa = 11.84RR187 pKa = 11.84KK188 pKa = 8.26ITSNSDD194 pKa = 2.74EE195 pKa = 4.4SLTINTTMGAIKK207 pKa = 10.0GQQLYY212 pKa = 8.43WVLEE216 pKa = 4.38IYY218 pKa = 10.62AQGQTNLMTGIAGTIDD234 pKa = 3.17TVVYY238 pKa = 10.24FKK240 pKa = 11.04DD241 pKa = 3.22AA242 pKa = 3.57
MM1 pKa = 7.83AYY3 pKa = 9.78HH4 pKa = 6.95RR5 pKa = 11.84KK6 pKa = 6.8TRR8 pKa = 11.84RR9 pKa = 11.84GHH11 pKa = 5.3RR12 pKa = 11.84SKK14 pKa = 11.08RR15 pKa = 11.84KK16 pKa = 6.49TPKK19 pKa = 10.05RR20 pKa = 11.84LSQHH24 pKa = 4.48MVRR27 pKa = 11.84AIKK30 pKa = 10.31AISQGEE36 pKa = 4.52VEE38 pKa = 4.71TKK40 pKa = 10.21RR41 pKa = 11.84YY42 pKa = 7.96MVSTDD47 pKa = 2.43ISTALTAAGYY57 pKa = 10.51VSGNNAVIRR66 pKa = 11.84RR67 pKa = 11.84NFLSDD72 pKa = 4.17LPRR75 pKa = 11.84EE76 pKa = 4.33SNTAVNSDD84 pKa = 2.96ASFIGDD90 pKa = 3.55QINLRR95 pKa = 11.84GLRR98 pKa = 11.84WSFHH102 pKa = 5.01GSILPTVNPDD112 pKa = 2.28AMYY115 pKa = 10.83RR116 pKa = 11.84FTVYY120 pKa = 10.27RR121 pKa = 11.84DD122 pKa = 3.14SFYY125 pKa = 10.9FAGTNGPGPGDD136 pKa = 3.79RR137 pKa = 11.84IFEE140 pKa = 4.3QQQSTIPTWSMWNTQVTDD158 pKa = 3.09IVFQRR163 pKa = 11.84KK164 pKa = 7.09FRR166 pKa = 11.84MRR168 pKa = 11.84VSSIGSEE175 pKa = 4.05VLDD178 pKa = 3.49KK179 pKa = 11.1KK180 pKa = 10.85FYY182 pKa = 10.18IPLRR186 pKa = 11.84RR187 pKa = 11.84KK188 pKa = 8.26ITSNSDD194 pKa = 2.74EE195 pKa = 4.4SLTINTTMGAIKK207 pKa = 10.0GQQLYY212 pKa = 8.43WVLEE216 pKa = 4.38IYY218 pKa = 10.62AQGQTNLMTGIAGTIDD234 pKa = 3.17TVVYY238 pKa = 10.24FKK240 pKa = 11.04DD241 pKa = 3.22AA242 pKa = 3.57
Molecular weight: 27.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
611 |
242 |
369 |
305.5 |
35.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.71 ± 0.317 | 0.982 ± 0.608 |
6.547 ± 0.984 | 5.237 ± 1.453 |
4.255 ± 0.18 | 5.892 ± 0.702 |
3.11 ± 0.902 | 6.219 ± 0.755 |
5.237 ± 0.173 | 6.383 ± 0.626 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.128 ± 0.73 | 4.583 ± 0.233 |
4.583 ± 0.791 | 3.601 ± 0.841 |
8.183 ± 0.306 | 6.547 ± 1.32 |
6.874 ± 1.373 | 6.547 ± 0.216 |
2.619 ± 0.598 | 3.764 ± 0.228 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
