
Pyrobaculum spherical virus (isolate United States/Yellowstone) (PSV)
Taxonomy: Viruses; Globuloviridae; Alphaglobulovirus; Alphaglobulovirus PSV; Pyrobaculum spherical virus
Average proteome isoelectric point is 7.61
Get precalculated fractions of proteins
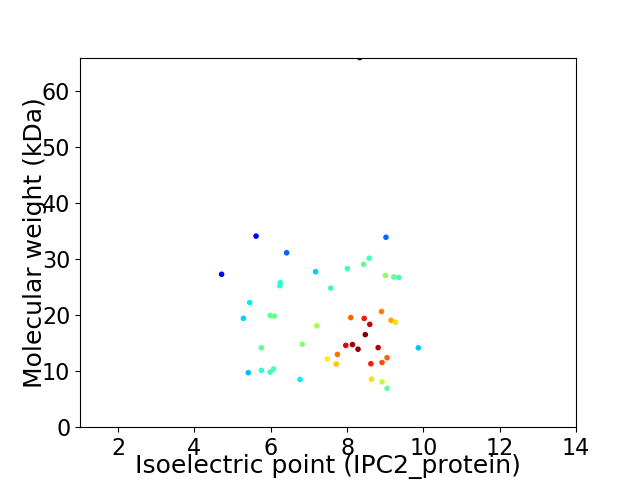
Virtual 2D-PAGE plot for 48 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
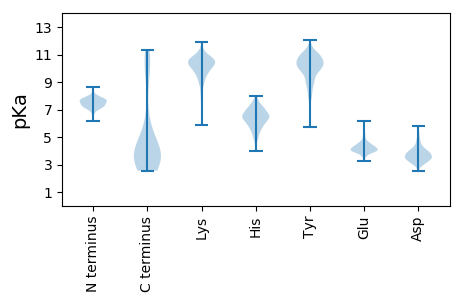
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q6ZYJ3|Q6ZYJ3_PSVY Uncharacterized protein OS=Pyrobaculum spherical virus (isolate United States/Yellowstone) OX=654907 PE=4 SV=1
MM1 pKa = 7.64SAFDD5 pKa = 3.95EE6 pKa = 4.68FNEE9 pKa = 4.47GFGLDD14 pKa = 3.49VSDD17 pKa = 4.38TPEE20 pKa = 3.66EE21 pKa = 3.93LAFEE25 pKa = 4.34TEE27 pKa = 4.26SAIEE31 pKa = 4.33EE32 pKa = 4.46IEE34 pKa = 4.47SEE36 pKa = 4.5TSPGDD41 pKa = 3.46QPKK44 pKa = 10.48GSEE47 pKa = 4.08PEE49 pKa = 4.29EE50 pKa = 3.62IRR52 pKa = 11.84VWAEE56 pKa = 3.41EE57 pKa = 4.06KK58 pKa = 10.43ARR60 pKa = 11.84KK61 pKa = 9.06AVEE64 pKa = 3.91EE65 pKa = 4.21GRR67 pKa = 11.84EE68 pKa = 3.99VTNWADD74 pKa = 3.35WIMGWRR80 pKa = 11.84TPNASEE86 pKa = 3.98KK87 pKa = 10.83KK88 pKa = 8.8MEE90 pKa = 3.85FMYY93 pKa = 9.74WYY95 pKa = 8.7TRR97 pKa = 11.84TYY99 pKa = 10.85LEE101 pKa = 3.94EE102 pKa = 5.17AKK104 pKa = 10.56DD105 pKa = 3.82IRR107 pKa = 11.84PDD109 pKa = 3.21IADD112 pKa = 3.56ALARR116 pKa = 11.84GMAGLAFGRR125 pKa = 11.84TDD127 pKa = 2.93WVASMLDD134 pKa = 3.7PQIMRR139 pKa = 11.84HH140 pKa = 6.16IYY142 pKa = 9.49TDD144 pKa = 3.55PEE146 pKa = 3.88VARR149 pKa = 11.84IYY151 pKa = 11.08SEE153 pKa = 3.87TRR155 pKa = 11.84DD156 pKa = 3.4MLRR159 pKa = 11.84RR160 pKa = 11.84VSDD163 pKa = 3.8YY164 pKa = 11.03YY165 pKa = 10.57ISLTTMEE172 pKa = 4.87LGKK175 pKa = 10.63VADD178 pKa = 5.08IIAEE182 pKa = 4.05AKK184 pKa = 10.59AKK186 pKa = 10.62GEE188 pKa = 3.98NPEE191 pKa = 3.92VVARR195 pKa = 11.84EE196 pKa = 3.66IAEE199 pKa = 4.12AVPRR203 pKa = 11.84LSPKK207 pKa = 10.13SLYY210 pKa = 10.53FNLYY214 pKa = 9.81YY215 pKa = 10.04IGRR218 pKa = 11.84SIGDD222 pKa = 3.44NYY224 pKa = 10.02VLEE227 pKa = 4.4VARR230 pKa = 11.84VLSKK234 pKa = 9.98MRR236 pKa = 11.84RR237 pKa = 11.84RR238 pKa = 3.76
MM1 pKa = 7.64SAFDD5 pKa = 3.95EE6 pKa = 4.68FNEE9 pKa = 4.47GFGLDD14 pKa = 3.49VSDD17 pKa = 4.38TPEE20 pKa = 3.66EE21 pKa = 3.93LAFEE25 pKa = 4.34TEE27 pKa = 4.26SAIEE31 pKa = 4.33EE32 pKa = 4.46IEE34 pKa = 4.47SEE36 pKa = 4.5TSPGDD41 pKa = 3.46QPKK44 pKa = 10.48GSEE47 pKa = 4.08PEE49 pKa = 4.29EE50 pKa = 3.62IRR52 pKa = 11.84VWAEE56 pKa = 3.41EE57 pKa = 4.06KK58 pKa = 10.43ARR60 pKa = 11.84KK61 pKa = 9.06AVEE64 pKa = 3.91EE65 pKa = 4.21GRR67 pKa = 11.84EE68 pKa = 3.99VTNWADD74 pKa = 3.35WIMGWRR80 pKa = 11.84TPNASEE86 pKa = 3.98KK87 pKa = 10.83KK88 pKa = 8.8MEE90 pKa = 3.85FMYY93 pKa = 9.74WYY95 pKa = 8.7TRR97 pKa = 11.84TYY99 pKa = 10.85LEE101 pKa = 3.94EE102 pKa = 5.17AKK104 pKa = 10.56DD105 pKa = 3.82IRR107 pKa = 11.84PDD109 pKa = 3.21IADD112 pKa = 3.56ALARR116 pKa = 11.84GMAGLAFGRR125 pKa = 11.84TDD127 pKa = 2.93WVASMLDD134 pKa = 3.7PQIMRR139 pKa = 11.84HH140 pKa = 6.16IYY142 pKa = 9.49TDD144 pKa = 3.55PEE146 pKa = 3.88VARR149 pKa = 11.84IYY151 pKa = 11.08SEE153 pKa = 3.87TRR155 pKa = 11.84DD156 pKa = 3.4MLRR159 pKa = 11.84RR160 pKa = 11.84VSDD163 pKa = 3.8YY164 pKa = 11.03YY165 pKa = 10.57ISLTTMEE172 pKa = 4.87LGKK175 pKa = 10.63VADD178 pKa = 5.08IIAEE182 pKa = 4.05AKK184 pKa = 10.59AKK186 pKa = 10.62GEE188 pKa = 3.98NPEE191 pKa = 3.92VVARR195 pKa = 11.84EE196 pKa = 3.66IAEE199 pKa = 4.12AVPRR203 pKa = 11.84LSPKK207 pKa = 10.13SLYY210 pKa = 10.53FNLYY214 pKa = 9.81YY215 pKa = 10.04IGRR218 pKa = 11.84SIGDD222 pKa = 3.44NYY224 pKa = 10.02VLEE227 pKa = 4.4VARR230 pKa = 11.84VLSKK234 pKa = 9.98MRR236 pKa = 11.84RR237 pKa = 11.84RR238 pKa = 3.76
Molecular weight: 27.32 kDa
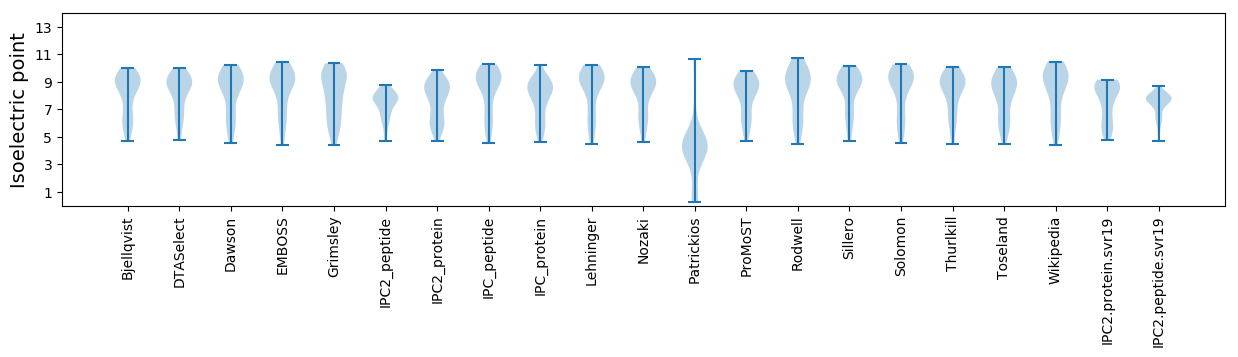
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6ZYF6|Q6ZYF6_PSVY Uncharacterized protein OS=Pyrobaculum spherical virus (isolate United States/Yellowstone) OX=654907 PE=1 SV=1
MM1 pKa = 7.21PRR3 pKa = 11.84KK4 pKa = 9.5MSDD7 pKa = 2.88TMHH10 pKa = 5.85TRR12 pKa = 11.84MIKK15 pKa = 7.32TLRR18 pKa = 11.84YY19 pKa = 8.27MITEE23 pKa = 4.76APCTTIARR31 pKa = 11.84VANKK35 pKa = 10.31LDD37 pKa = 3.97VSWSMARR44 pKa = 11.84NALYY48 pKa = 10.32QLAKK52 pKa = 10.69NKK54 pKa = 9.16MVLKK58 pKa = 10.6VEE60 pKa = 4.5ISRR63 pKa = 11.84ATIWCVDD70 pKa = 3.26EE71 pKa = 4.84NAATNAIFEE80 pKa = 4.65IKK82 pKa = 8.82RR83 pKa = 11.84TLWRR87 pKa = 11.84LICNSRR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84YY96 pKa = 7.93ITPSDD101 pKa = 3.8MLRR104 pKa = 11.84LIAADD109 pKa = 3.91TEE111 pKa = 4.17AHH113 pKa = 6.46NIFAKK118 pKa = 10.68YY119 pKa = 10.23VDD121 pKa = 3.76VNSTHH126 pKa = 5.72GHH128 pKa = 4.75TLHH131 pKa = 7.09FIASALEE138 pKa = 4.1DD139 pKa = 3.96LLGKK143 pKa = 9.87PMDD146 pKa = 3.4KK147 pKa = 10.22RR148 pKa = 11.84QRR150 pKa = 11.84RR151 pKa = 11.84LLFHH155 pKa = 5.46VTPRR159 pKa = 11.84FCKK162 pKa = 10.11KK163 pKa = 10.52APTLADD169 pKa = 3.31INLSHH174 pKa = 6.09YY175 pKa = 8.63TSAYY179 pKa = 9.68RR180 pKa = 11.84LVTFKK185 pKa = 11.22VPTAMHH191 pKa = 6.82NDD193 pKa = 3.2MLEE196 pKa = 3.98AARR199 pKa = 11.84TLGLTTSEE207 pKa = 4.53LVTMAIDD214 pKa = 3.58RR215 pKa = 11.84LLSQYY220 pKa = 10.91RR221 pKa = 11.84HH222 pKa = 5.52VLDD225 pKa = 4.36GKK227 pKa = 11.06GGADD231 pKa = 3.85LKK233 pKa = 11.63ANN235 pKa = 3.78
MM1 pKa = 7.21PRR3 pKa = 11.84KK4 pKa = 9.5MSDD7 pKa = 2.88TMHH10 pKa = 5.85TRR12 pKa = 11.84MIKK15 pKa = 7.32TLRR18 pKa = 11.84YY19 pKa = 8.27MITEE23 pKa = 4.76APCTTIARR31 pKa = 11.84VANKK35 pKa = 10.31LDD37 pKa = 3.97VSWSMARR44 pKa = 11.84NALYY48 pKa = 10.32QLAKK52 pKa = 10.69NKK54 pKa = 9.16MVLKK58 pKa = 10.6VEE60 pKa = 4.5ISRR63 pKa = 11.84ATIWCVDD70 pKa = 3.26EE71 pKa = 4.84NAATNAIFEE80 pKa = 4.65IKK82 pKa = 8.82RR83 pKa = 11.84TLWRR87 pKa = 11.84LICNSRR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84YY96 pKa = 7.93ITPSDD101 pKa = 3.8MLRR104 pKa = 11.84LIAADD109 pKa = 3.91TEE111 pKa = 4.17AHH113 pKa = 6.46NIFAKK118 pKa = 10.68YY119 pKa = 10.23VDD121 pKa = 3.76VNSTHH126 pKa = 5.72GHH128 pKa = 4.75TLHH131 pKa = 7.09FIASALEE138 pKa = 4.1DD139 pKa = 3.96LLGKK143 pKa = 9.87PMDD146 pKa = 3.4KK147 pKa = 10.22RR148 pKa = 11.84QRR150 pKa = 11.84RR151 pKa = 11.84LLFHH155 pKa = 5.46VTPRR159 pKa = 11.84FCKK162 pKa = 10.11KK163 pKa = 10.52APTLADD169 pKa = 3.31INLSHH174 pKa = 6.09YY175 pKa = 8.63TSAYY179 pKa = 9.68RR180 pKa = 11.84LVTFKK185 pKa = 11.22VPTAMHH191 pKa = 6.82NDD193 pKa = 3.2MLEE196 pKa = 3.98AARR199 pKa = 11.84TLGLTTSEE207 pKa = 4.53LVTMAIDD214 pKa = 3.58RR215 pKa = 11.84LLSQYY220 pKa = 10.91RR221 pKa = 11.84HH222 pKa = 5.52VLDD225 pKa = 4.36GKK227 pKa = 11.06GGADD231 pKa = 3.85LKK233 pKa = 11.63ANN235 pKa = 3.78
Molecular weight: 26.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8428 |
61 |
581 |
175.6 |
19.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.792 ± 0.469 | 1.009 ± 0.161 |
4.153 ± 0.302 | 5.019 ± 0.433 |
4.117 ± 0.301 | 7.202 ± 0.42 |
1.661 ± 0.159 | 6.739 ± 0.261 |
5.233 ± 0.338 | 10.168 ± 0.535 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.919 ± 0.169 | 3.429 ± 0.18 |
4.224 ± 0.293 | 2.219 ± 0.21 |
5.327 ± 0.361 | 5.256 ± 0.328 |
5.672 ± 0.418 | 9.955 ± 0.367 |
1.614 ± 0.168 | 5.244 ± 0.346 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
