
Candida maltosa (strain Xu316) (Yeast)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Debaryomycetaceae; Candida/Lodderomyces clade; Candida; Candida maltosa
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
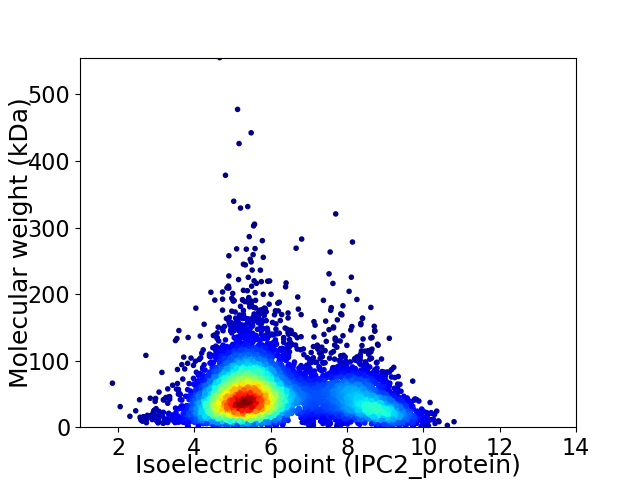
Virtual 2D-PAGE plot for 5976 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M3HFY7|M3HFY7_CANMX Alpha-1 2-mannosyltransferase putative OS=Candida maltosa (strain Xu316) OX=1245528 GN=G210_3625 PE=3 SV=1
MM1 pKa = 7.67IFLLLLSLIVATVVEE16 pKa = 4.88CSLKK20 pKa = 10.12IDD22 pKa = 4.1FEE24 pKa = 4.46VHH26 pKa = 5.6KK27 pKa = 11.05ASFNPHH33 pKa = 4.6LMKK36 pKa = 10.56RR37 pKa = 11.84SSPVLSIINNKK48 pKa = 8.93SLYY51 pKa = 8.29ITSLAIGSNKK61 pKa = 10.18DD62 pKa = 3.78PISVSIDD69 pKa = 3.06TGSADD74 pKa = 3.66LWVMGSDD81 pKa = 3.54VTCFDD86 pKa = 3.4TSEE89 pKa = 4.25LHH91 pKa = 6.73VDD93 pKa = 4.44GAPSLPEE100 pKa = 3.88IFDD103 pKa = 5.56DD104 pKa = 4.31INPDD108 pKa = 3.53YY109 pKa = 11.38SCTANGTFDD118 pKa = 3.73TANSTTFKK126 pKa = 10.4STDD129 pKa = 3.25DD130 pKa = 3.69NFVIGYY136 pKa = 7.95TDD138 pKa = 3.76GSAAIGKK145 pKa = 8.59WGIDD149 pKa = 3.01SVQFGNSTINEE160 pKa = 4.12LRR162 pKa = 11.84LGIASQSSVSDD173 pKa = 4.41GILGIGIPDD182 pKa = 4.38GYY184 pKa = 11.16DD185 pKa = 3.0NFPIQLKK192 pKa = 7.92NQKK195 pKa = 10.38LIDD198 pKa = 3.56KK199 pKa = 9.23VAYY202 pKa = 9.47SVYY205 pKa = 10.45LNSSDD210 pKa = 4.96AIGGTVLFGAIDD222 pKa = 3.4HH223 pKa = 6.69AKK225 pKa = 10.67YY226 pKa = 10.27EE227 pKa = 4.44GALTTVPLTSDD238 pKa = 3.18TLLSVNVTYY247 pKa = 11.01EE248 pKa = 3.87NVNYY252 pKa = 10.6SVILDD257 pKa = 3.67TGSTFSIFPDD267 pKa = 2.89TWINDD272 pKa = 3.17FGTLLNGTYY281 pKa = 10.49DD282 pKa = 3.82DD283 pKa = 4.01NEE285 pKa = 3.79EE286 pKa = 4.35VYY288 pKa = 10.67RR289 pKa = 11.84IDD291 pKa = 4.36CDD293 pKa = 3.7SRR295 pKa = 11.84DD296 pKa = 3.61DD297 pKa = 3.82FFEE300 pKa = 4.14FEE302 pKa = 4.22IGNASFAIPVEE313 pKa = 4.08DD314 pKa = 5.48FIVEE318 pKa = 4.02NDD320 pKa = 3.86DD321 pKa = 3.44VCYY324 pKa = 9.92LAIMGNSVIGGDD336 pKa = 3.96GILFGGDD343 pKa = 2.99ILRR346 pKa = 11.84SIYY349 pKa = 10.45LVYY352 pKa = 10.7DD353 pKa = 3.67LEE355 pKa = 5.2DD356 pKa = 3.39RR357 pKa = 11.84TISVAPVRR365 pKa = 11.84YY366 pKa = 8.27TDD368 pKa = 4.72DD369 pKa = 3.52EE370 pKa = 5.19DD371 pKa = 4.76IEE373 pKa = 4.37EE374 pKa = 4.98LGNQTVAVDD383 pKa = 3.55NSTVSTSQTSVLPTSQTSALSSTSSVNQARR413 pKa = 11.84VIGSFTVSTLMGFILTFII431 pKa = 4.55
MM1 pKa = 7.67IFLLLLSLIVATVVEE16 pKa = 4.88CSLKK20 pKa = 10.12IDD22 pKa = 4.1FEE24 pKa = 4.46VHH26 pKa = 5.6KK27 pKa = 11.05ASFNPHH33 pKa = 4.6LMKK36 pKa = 10.56RR37 pKa = 11.84SSPVLSIINNKK48 pKa = 8.93SLYY51 pKa = 8.29ITSLAIGSNKK61 pKa = 10.18DD62 pKa = 3.78PISVSIDD69 pKa = 3.06TGSADD74 pKa = 3.66LWVMGSDD81 pKa = 3.54VTCFDD86 pKa = 3.4TSEE89 pKa = 4.25LHH91 pKa = 6.73VDD93 pKa = 4.44GAPSLPEE100 pKa = 3.88IFDD103 pKa = 5.56DD104 pKa = 4.31INPDD108 pKa = 3.53YY109 pKa = 11.38SCTANGTFDD118 pKa = 3.73TANSTTFKK126 pKa = 10.4STDD129 pKa = 3.25DD130 pKa = 3.69NFVIGYY136 pKa = 7.95TDD138 pKa = 3.76GSAAIGKK145 pKa = 8.59WGIDD149 pKa = 3.01SVQFGNSTINEE160 pKa = 4.12LRR162 pKa = 11.84LGIASQSSVSDD173 pKa = 4.41GILGIGIPDD182 pKa = 4.38GYY184 pKa = 11.16DD185 pKa = 3.0NFPIQLKK192 pKa = 7.92NQKK195 pKa = 10.38LIDD198 pKa = 3.56KK199 pKa = 9.23VAYY202 pKa = 9.47SVYY205 pKa = 10.45LNSSDD210 pKa = 4.96AIGGTVLFGAIDD222 pKa = 3.4HH223 pKa = 6.69AKK225 pKa = 10.67YY226 pKa = 10.27EE227 pKa = 4.44GALTTVPLTSDD238 pKa = 3.18TLLSVNVTYY247 pKa = 11.01EE248 pKa = 3.87NVNYY252 pKa = 10.6SVILDD257 pKa = 3.67TGSTFSIFPDD267 pKa = 2.89TWINDD272 pKa = 3.17FGTLLNGTYY281 pKa = 10.49DD282 pKa = 3.82DD283 pKa = 4.01NEE285 pKa = 3.79EE286 pKa = 4.35VYY288 pKa = 10.67RR289 pKa = 11.84IDD291 pKa = 4.36CDD293 pKa = 3.7SRR295 pKa = 11.84DD296 pKa = 3.61DD297 pKa = 3.82FFEE300 pKa = 4.14FEE302 pKa = 4.22IGNASFAIPVEE313 pKa = 4.08DD314 pKa = 5.48FIVEE318 pKa = 4.02NDD320 pKa = 3.86DD321 pKa = 3.44VCYY324 pKa = 9.92LAIMGNSVIGGDD336 pKa = 3.96GILFGGDD343 pKa = 2.99ILRR346 pKa = 11.84SIYY349 pKa = 10.45LVYY352 pKa = 10.7DD353 pKa = 3.67LEE355 pKa = 5.2DD356 pKa = 3.39RR357 pKa = 11.84TISVAPVRR365 pKa = 11.84YY366 pKa = 8.27TDD368 pKa = 4.72DD369 pKa = 3.52EE370 pKa = 5.19DD371 pKa = 4.76IEE373 pKa = 4.37EE374 pKa = 4.98LGNQTVAVDD383 pKa = 3.55NSTVSTSQTSVLPTSQTSALSSTSSVNQARR413 pKa = 11.84VIGSFTVSTLMGFILTFII431 pKa = 4.55
Molecular weight: 46.52 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M3JVG2|M3JVG2_CANMX Elongin-A OS=Candida maltosa (strain Xu316) OX=1245528 GN=G210_2836 PE=4 SV=1
SS1 pKa = 5.96TRR3 pKa = 11.84ATSCTTTTNSSSTVASSGSANTSTRR28 pKa = 11.84VTRR31 pKa = 11.84SRR33 pKa = 11.84TTTRR37 pKa = 11.84STTARR42 pKa = 11.84KK43 pKa = 8.19STTVSTILPPVSSGPLKK60 pKa = 10.93NITNTVSARR69 pKa = 11.84SRR71 pKa = 11.84RR72 pKa = 11.84VKK74 pKa = 9.58TASKK78 pKa = 10.41KK79 pKa = 10.15
SS1 pKa = 5.96TRR3 pKa = 11.84ATSCTTTTNSSSTVASSGSANTSTRR28 pKa = 11.84VTRR31 pKa = 11.84SRR33 pKa = 11.84TTTRR37 pKa = 11.84STTARR42 pKa = 11.84KK43 pKa = 8.19STTVSTILPPVSSGPLKK60 pKa = 10.93NITNTVSARR69 pKa = 11.84SRR71 pKa = 11.84RR72 pKa = 11.84VKK74 pKa = 9.58TASKK78 pKa = 10.41KK79 pKa = 10.15
Molecular weight: 8.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2856021 |
10 |
4888 |
477.9 |
53.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.178 ± 0.031 | 1.057 ± 0.012 |
6.147 ± 0.027 | 6.722 ± 0.035 |
4.504 ± 0.022 | 5.125 ± 0.03 |
2.089 ± 0.014 | 6.886 ± 0.025 |
7.27 ± 0.033 | 9.182 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.823 ± 0.011 | 6.178 ± 0.029 |
4.697 ± 0.03 | 4.258 ± 0.032 |
3.792 ± 0.02 | 8.701 ± 0.042 |
6.082 ± 0.032 | 5.772 ± 0.025 |
0.99 ± 0.011 | 3.549 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
