
BtMr-AlphaCoV/SAX2011
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; Myotacovirus; Myotis ricketti alphacoronavirus Sax-2011
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
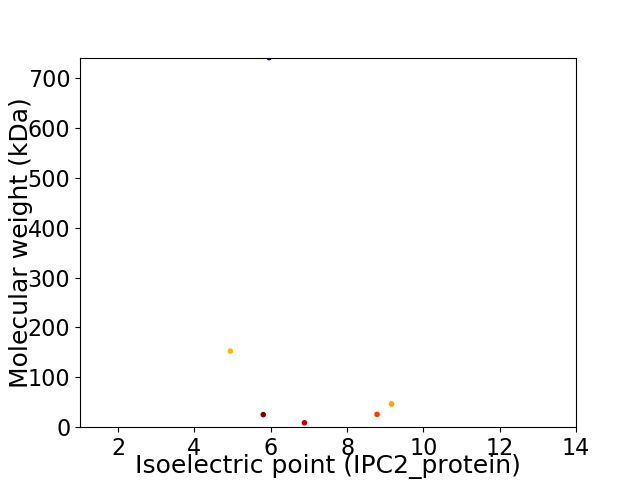
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
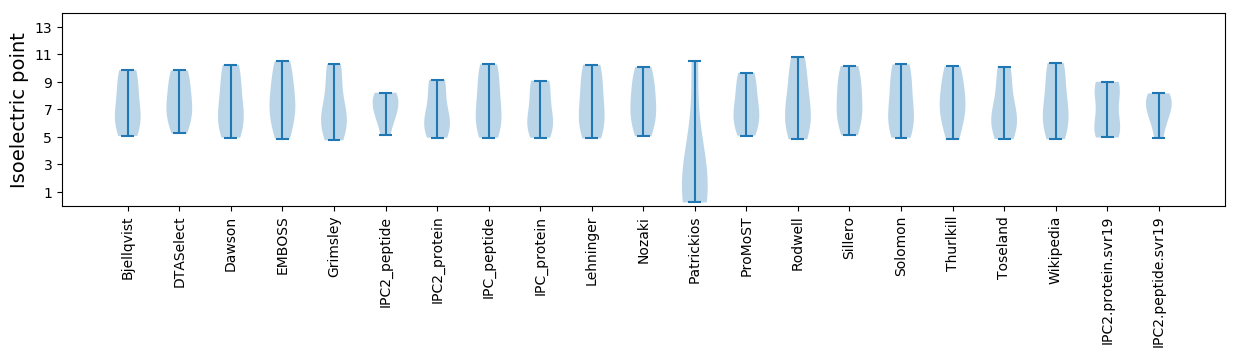
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U1WHE1|A0A0U1WHE1_9ALPC Envelope small membrane protein OS=BtMr-AlphaCoV/SAX2011 OX=1503289 GN=E PE=3 SV=1
MM1 pKa = 6.69WHH3 pKa = 6.48VIVALVFIPFLSAANTGTCSNTINRR28 pKa = 11.84FMYY31 pKa = 10.38LKK33 pKa = 10.62LGLPVPTNDD42 pKa = 4.41AYY44 pKa = 10.35VTGYY48 pKa = 11.02LPIPSNWTCSDD59 pKa = 4.21GYY61 pKa = 9.12WQYY64 pKa = 12.08KK65 pKa = 7.89NAHH68 pKa = 5.89GLFVSYY74 pKa = 10.64HH75 pKa = 4.55STALDD80 pKa = 3.41FAIGVSSSTSYY91 pKa = 10.68AEE93 pKa = 3.66KK94 pKa = 9.31WGLYY98 pKa = 8.7FWQKK102 pKa = 10.31NNEE105 pKa = 3.48GRR107 pKa = 11.84AVFRR111 pKa = 11.84ICKK114 pKa = 8.79WDD116 pKa = 3.68ASAQTNFAPDD126 pKa = 2.69RR127 pKa = 11.84HH128 pKa = 5.12TARR131 pKa = 11.84DD132 pKa = 3.77CFVNKK137 pKa = 9.76QIPMVFKK144 pKa = 10.4HH145 pKa = 5.73QGNEE149 pKa = 3.64IVGVSWSGSRR159 pKa = 11.84VTLHH163 pKa = 5.65TLDD166 pKa = 3.7RR167 pKa = 11.84VFSFIVEE174 pKa = 4.39GAEE177 pKa = 3.75SWDD180 pKa = 3.79YY181 pKa = 11.12ATLRR185 pKa = 11.84CANGYY190 pKa = 9.72ACAHH194 pKa = 4.46QVVYY198 pKa = 11.2NPVTVIVTTVAAGIQKK214 pKa = 8.21YY215 pKa = 6.43TLCDD219 pKa = 3.16NCTGFPQHH227 pKa = 6.21VFATMEE233 pKa = 4.2NGEE236 pKa = 4.64IPPSFNFANWFYY248 pKa = 9.91LTNSSSPVSSRR259 pKa = 11.84VVGLQPLLLTCLWPIPALLGTATDD283 pKa = 3.25ITFDD287 pKa = 3.69RR288 pKa = 11.84NGTSDD293 pKa = 3.36VRR295 pKa = 11.84CNGFASNEE303 pKa = 3.99TADD306 pKa = 3.97AMRR309 pKa = 11.84FSLNFTDD316 pKa = 3.72SAVFAKK322 pKa = 10.53EE323 pKa = 3.72GVITLKK329 pKa = 10.01TLSNTFKK336 pKa = 10.67FSCSNSSTYY345 pKa = 8.85QAPYY349 pKa = 9.92VIPFGHH355 pKa = 7.2IDD357 pKa = 3.31QPYY360 pKa = 7.7YY361 pKa = 11.08CFTTFYY367 pKa = 10.53INEE370 pKa = 4.32TAGTTTTSFVGMLPPVVRR388 pKa = 11.84EE389 pKa = 3.81FVITKK394 pKa = 8.85TGNVYY399 pKa = 10.87LNGYY403 pKa = 10.02RR404 pKa = 11.84IFTVDD409 pKa = 4.2DD410 pKa = 3.9VVSVNFNISSTDD422 pKa = 3.08HH423 pKa = 6.65RR424 pKa = 11.84DD425 pKa = 2.95FWTVAFVKK433 pKa = 9.4NTEE436 pKa = 3.87VMLDD440 pKa = 3.4IEE442 pKa = 4.38DD443 pKa = 4.27TYY445 pKa = 11.24IKK447 pKa = 10.42QLLYY451 pKa = 11.09CNTPLNVVKK460 pKa = 10.35CQQLKK465 pKa = 10.06FVLDD469 pKa = 4.07DD470 pKa = 3.68GFYY473 pKa = 10.29SYY475 pKa = 11.27SSPVDD480 pKa = 3.23EE481 pKa = 4.52VLPRR485 pKa = 11.84TIVRR489 pKa = 11.84LPRR492 pKa = 11.84LMTHH496 pKa = 6.24NFLNFTIFVSFYY508 pKa = 10.9FDD510 pKa = 3.93DD511 pKa = 5.11DD512 pKa = 3.61KK513 pKa = 11.39QARR516 pKa = 11.84PDD518 pKa = 3.35GGFYY522 pKa = 10.7EE523 pKa = 5.23CATCAPKK530 pKa = 10.45YY531 pKa = 10.09YY532 pKa = 10.72KK533 pKa = 10.56LAFVDD538 pKa = 4.16DD539 pKa = 3.74WTSAVSSNVTSICVNYY555 pKa = 10.79ASFTTRR561 pKa = 11.84LFTFYY566 pKa = 10.84AGTTAGVHH574 pKa = 6.28LGLEE578 pKa = 4.41TGTCPFSFDD587 pKa = 3.4TLNNYY592 pKa = 7.65LTFGSLCFSLVANGGCTMNIVTQGPYY618 pKa = 10.77GLPHH622 pKa = 7.24TIAVLYY628 pKa = 10.27VSYY631 pKa = 11.01TEE633 pKa = 4.24GDD635 pKa = 3.78NIIGVPLSNIPPLGVSDD652 pKa = 4.0MSQVYY657 pKa = 10.77LDD659 pKa = 3.37TCTTYY664 pKa = 10.4TIYY667 pKa = 11.16GMTGRR672 pKa = 11.84GVITRR677 pKa = 11.84SNNTFITGLYY687 pKa = 6.7YY688 pKa = 10.52TSNAGNLLAYY698 pKa = 9.62KK699 pKa = 10.43NSTTGVVYY707 pKa = 10.5NVYY710 pKa = 9.9PCQLSSQVAVISDD723 pKa = 4.98AIVGMASSTPNVSIDD738 pKa = 3.61FNVTVVADD746 pKa = 3.52NFYY749 pKa = 10.78YY750 pKa = 10.73LSNSAQPCDD759 pKa = 3.98QPVLTYY765 pKa = 10.79AGIGICSDD773 pKa = 3.38GSITNSTARR782 pKa = 11.84RR783 pKa = 11.84AAADD787 pKa = 3.49PVSPVISGNISVPTNFTFSVQVEE810 pKa = 4.48YY811 pKa = 10.39IQLMLKK817 pKa = 10.04PVTVDD822 pKa = 2.84CSVYY826 pKa = 10.32VCNGNPRR833 pKa = 11.84CLQLLAQYY841 pKa = 11.14ASACRR846 pKa = 11.84TIEE849 pKa = 3.99QALQLSARR857 pKa = 11.84LEE859 pKa = 4.32SVEE862 pKa = 4.2VNSMISISQEE872 pKa = 3.4ALALGVIDD880 pKa = 4.95NFKK883 pKa = 10.97HH884 pKa = 6.74DD885 pKa = 4.11FNLTNVLPASVGAKK899 pKa = 9.92SAVEE903 pKa = 4.48DD904 pKa = 3.82LLFDD908 pKa = 3.81KK909 pKa = 11.09VVTSGLGTVDD919 pKa = 4.34ADD921 pKa = 3.94YY922 pKa = 11.12KK923 pKa = 10.27EE924 pKa = 4.6CASRR928 pKa = 11.84TANTVAEE935 pKa = 4.59VGCVQYY941 pKa = 11.44YY942 pKa = 9.74NGIMVLPGVVDD953 pKa = 3.61QSLLAQYY960 pKa = 10.48SAALTGAMVFGGVTAGAAVPFSIAVQSRR988 pKa = 11.84LNYY991 pKa = 10.06LALQTDD997 pKa = 4.07VLQRR1001 pKa = 11.84NQQQLANSFNAAMGNITEE1019 pKa = 4.28AFGRR1023 pKa = 11.84VNDD1026 pKa = 5.09AIEE1029 pKa = 4.21QTSHH1033 pKa = 7.28AISTVAQALDD1043 pKa = 3.45KK1044 pKa = 11.12VQTVVNDD1051 pKa = 3.14QGLALSQLTKK1061 pKa = 10.65QLASNFQAISSSIEE1075 pKa = 3.69DD1076 pKa = 3.87LYY1078 pKa = 11.76NRR1080 pKa = 11.84LDD1082 pKa = 3.53RR1083 pKa = 11.84VEE1085 pKa = 5.91ADD1087 pKa = 3.61LQVDD1091 pKa = 3.85RR1092 pKa = 11.84LITGRR1097 pKa = 11.84LAALNAFVAQQLTKK1111 pKa = 9.35YY1112 pKa = 8.42TDD1114 pKa = 3.16VRR1116 pKa = 11.84ASRR1119 pKa = 11.84QLAQDD1124 pKa = 4.7KK1125 pKa = 10.87INEE1128 pKa = 4.46CVKK1131 pKa = 10.55SQSFRR1136 pKa = 11.84YY1137 pKa = 8.66GFCGNGTHH1145 pKa = 6.04VFSVVNAAPDD1155 pKa = 3.22GMMFFHH1161 pKa = 7.24SVLLPTAYY1169 pKa = 9.73MEE1171 pKa = 4.19VAAFSGLCVEE1181 pKa = 4.56GNGYY1185 pKa = 7.66VLRR1188 pKa = 11.84DD1189 pKa = 3.41TGNVLFEE1196 pKa = 4.9KK1197 pKa = 10.4NGQYY1201 pKa = 10.86LITARR1206 pKa = 11.84KK1207 pKa = 7.74MFEE1210 pKa = 3.7PRR1212 pKa = 11.84VPQTSDD1218 pKa = 2.95FVQITGCDD1226 pKa = 3.24VVYY1229 pKa = 11.09LNVTRR1234 pKa = 11.84DD1235 pKa = 3.69EE1236 pKa = 4.88LPTVIPDD1243 pKa = 3.98YY1244 pKa = 10.97IDD1246 pKa = 3.46VNSTVEE1252 pKa = 4.81DD1253 pKa = 3.67ILSKK1257 pKa = 10.79LPNRR1261 pKa = 11.84TTPEE1265 pKa = 3.49FDD1267 pKa = 4.23LDD1269 pKa = 3.82IFNATYY1275 pKa = 11.19LNLTGEE1281 pKa = 4.41IADD1284 pKa = 3.56LTARR1288 pKa = 11.84SEE1290 pKa = 4.34SLKK1293 pKa = 9.87NTTLEE1298 pKa = 4.05LKK1300 pKa = 10.58EE1301 pKa = 4.49LIANINATLVDD1312 pKa = 4.66LEE1314 pKa = 4.13WLNRR1318 pKa = 11.84VEE1320 pKa = 5.97TYY1322 pKa = 10.41IKK1324 pKa = 9.23WPWWVWLIIVLVLILFTCLMLFCCCSTGCCGIFSCMASSCGACCDD1369 pKa = 3.2IRR1371 pKa = 11.84GTKK1374 pKa = 8.05LQRR1377 pKa = 11.84YY1378 pKa = 5.97EE1379 pKa = 4.8AIEE1382 pKa = 4.12KK1383 pKa = 10.11VHH1385 pKa = 5.3VQQ1387 pKa = 3.06
MM1 pKa = 6.69WHH3 pKa = 6.48VIVALVFIPFLSAANTGTCSNTINRR28 pKa = 11.84FMYY31 pKa = 10.38LKK33 pKa = 10.62LGLPVPTNDD42 pKa = 4.41AYY44 pKa = 10.35VTGYY48 pKa = 11.02LPIPSNWTCSDD59 pKa = 4.21GYY61 pKa = 9.12WQYY64 pKa = 12.08KK65 pKa = 7.89NAHH68 pKa = 5.89GLFVSYY74 pKa = 10.64HH75 pKa = 4.55STALDD80 pKa = 3.41FAIGVSSSTSYY91 pKa = 10.68AEE93 pKa = 3.66KK94 pKa = 9.31WGLYY98 pKa = 8.7FWQKK102 pKa = 10.31NNEE105 pKa = 3.48GRR107 pKa = 11.84AVFRR111 pKa = 11.84ICKK114 pKa = 8.79WDD116 pKa = 3.68ASAQTNFAPDD126 pKa = 2.69RR127 pKa = 11.84HH128 pKa = 5.12TARR131 pKa = 11.84DD132 pKa = 3.77CFVNKK137 pKa = 9.76QIPMVFKK144 pKa = 10.4HH145 pKa = 5.73QGNEE149 pKa = 3.64IVGVSWSGSRR159 pKa = 11.84VTLHH163 pKa = 5.65TLDD166 pKa = 3.7RR167 pKa = 11.84VFSFIVEE174 pKa = 4.39GAEE177 pKa = 3.75SWDD180 pKa = 3.79YY181 pKa = 11.12ATLRR185 pKa = 11.84CANGYY190 pKa = 9.72ACAHH194 pKa = 4.46QVVYY198 pKa = 11.2NPVTVIVTTVAAGIQKK214 pKa = 8.21YY215 pKa = 6.43TLCDD219 pKa = 3.16NCTGFPQHH227 pKa = 6.21VFATMEE233 pKa = 4.2NGEE236 pKa = 4.64IPPSFNFANWFYY248 pKa = 9.91LTNSSSPVSSRR259 pKa = 11.84VVGLQPLLLTCLWPIPALLGTATDD283 pKa = 3.25ITFDD287 pKa = 3.69RR288 pKa = 11.84NGTSDD293 pKa = 3.36VRR295 pKa = 11.84CNGFASNEE303 pKa = 3.99TADD306 pKa = 3.97AMRR309 pKa = 11.84FSLNFTDD316 pKa = 3.72SAVFAKK322 pKa = 10.53EE323 pKa = 3.72GVITLKK329 pKa = 10.01TLSNTFKK336 pKa = 10.67FSCSNSSTYY345 pKa = 8.85QAPYY349 pKa = 9.92VIPFGHH355 pKa = 7.2IDD357 pKa = 3.31QPYY360 pKa = 7.7YY361 pKa = 11.08CFTTFYY367 pKa = 10.53INEE370 pKa = 4.32TAGTTTTSFVGMLPPVVRR388 pKa = 11.84EE389 pKa = 3.81FVITKK394 pKa = 8.85TGNVYY399 pKa = 10.87LNGYY403 pKa = 10.02RR404 pKa = 11.84IFTVDD409 pKa = 4.2DD410 pKa = 3.9VVSVNFNISSTDD422 pKa = 3.08HH423 pKa = 6.65RR424 pKa = 11.84DD425 pKa = 2.95FWTVAFVKK433 pKa = 9.4NTEE436 pKa = 3.87VMLDD440 pKa = 3.4IEE442 pKa = 4.38DD443 pKa = 4.27TYY445 pKa = 11.24IKK447 pKa = 10.42QLLYY451 pKa = 11.09CNTPLNVVKK460 pKa = 10.35CQQLKK465 pKa = 10.06FVLDD469 pKa = 4.07DD470 pKa = 3.68GFYY473 pKa = 10.29SYY475 pKa = 11.27SSPVDD480 pKa = 3.23EE481 pKa = 4.52VLPRR485 pKa = 11.84TIVRR489 pKa = 11.84LPRR492 pKa = 11.84LMTHH496 pKa = 6.24NFLNFTIFVSFYY508 pKa = 10.9FDD510 pKa = 3.93DD511 pKa = 5.11DD512 pKa = 3.61KK513 pKa = 11.39QARR516 pKa = 11.84PDD518 pKa = 3.35GGFYY522 pKa = 10.7EE523 pKa = 5.23CATCAPKK530 pKa = 10.45YY531 pKa = 10.09YY532 pKa = 10.72KK533 pKa = 10.56LAFVDD538 pKa = 4.16DD539 pKa = 3.74WTSAVSSNVTSICVNYY555 pKa = 10.79ASFTTRR561 pKa = 11.84LFTFYY566 pKa = 10.84AGTTAGVHH574 pKa = 6.28LGLEE578 pKa = 4.41TGTCPFSFDD587 pKa = 3.4TLNNYY592 pKa = 7.65LTFGSLCFSLVANGGCTMNIVTQGPYY618 pKa = 10.77GLPHH622 pKa = 7.24TIAVLYY628 pKa = 10.27VSYY631 pKa = 11.01TEE633 pKa = 4.24GDD635 pKa = 3.78NIIGVPLSNIPPLGVSDD652 pKa = 4.0MSQVYY657 pKa = 10.77LDD659 pKa = 3.37TCTTYY664 pKa = 10.4TIYY667 pKa = 11.16GMTGRR672 pKa = 11.84GVITRR677 pKa = 11.84SNNTFITGLYY687 pKa = 6.7YY688 pKa = 10.52TSNAGNLLAYY698 pKa = 9.62KK699 pKa = 10.43NSTTGVVYY707 pKa = 10.5NVYY710 pKa = 9.9PCQLSSQVAVISDD723 pKa = 4.98AIVGMASSTPNVSIDD738 pKa = 3.61FNVTVVADD746 pKa = 3.52NFYY749 pKa = 10.78YY750 pKa = 10.73LSNSAQPCDD759 pKa = 3.98QPVLTYY765 pKa = 10.79AGIGICSDD773 pKa = 3.38GSITNSTARR782 pKa = 11.84RR783 pKa = 11.84AAADD787 pKa = 3.49PVSPVISGNISVPTNFTFSVQVEE810 pKa = 4.48YY811 pKa = 10.39IQLMLKK817 pKa = 10.04PVTVDD822 pKa = 2.84CSVYY826 pKa = 10.32VCNGNPRR833 pKa = 11.84CLQLLAQYY841 pKa = 11.14ASACRR846 pKa = 11.84TIEE849 pKa = 3.99QALQLSARR857 pKa = 11.84LEE859 pKa = 4.32SVEE862 pKa = 4.2VNSMISISQEE872 pKa = 3.4ALALGVIDD880 pKa = 4.95NFKK883 pKa = 10.97HH884 pKa = 6.74DD885 pKa = 4.11FNLTNVLPASVGAKK899 pKa = 9.92SAVEE903 pKa = 4.48DD904 pKa = 3.82LLFDD908 pKa = 3.81KK909 pKa = 11.09VVTSGLGTVDD919 pKa = 4.34ADD921 pKa = 3.94YY922 pKa = 11.12KK923 pKa = 10.27EE924 pKa = 4.6CASRR928 pKa = 11.84TANTVAEE935 pKa = 4.59VGCVQYY941 pKa = 11.44YY942 pKa = 9.74NGIMVLPGVVDD953 pKa = 3.61QSLLAQYY960 pKa = 10.48SAALTGAMVFGGVTAGAAVPFSIAVQSRR988 pKa = 11.84LNYY991 pKa = 10.06LALQTDD997 pKa = 4.07VLQRR1001 pKa = 11.84NQQQLANSFNAAMGNITEE1019 pKa = 4.28AFGRR1023 pKa = 11.84VNDD1026 pKa = 5.09AIEE1029 pKa = 4.21QTSHH1033 pKa = 7.28AISTVAQALDD1043 pKa = 3.45KK1044 pKa = 11.12VQTVVNDD1051 pKa = 3.14QGLALSQLTKK1061 pKa = 10.65QLASNFQAISSSIEE1075 pKa = 3.69DD1076 pKa = 3.87LYY1078 pKa = 11.76NRR1080 pKa = 11.84LDD1082 pKa = 3.53RR1083 pKa = 11.84VEE1085 pKa = 5.91ADD1087 pKa = 3.61LQVDD1091 pKa = 3.85RR1092 pKa = 11.84LITGRR1097 pKa = 11.84LAALNAFVAQQLTKK1111 pKa = 9.35YY1112 pKa = 8.42TDD1114 pKa = 3.16VRR1116 pKa = 11.84ASRR1119 pKa = 11.84QLAQDD1124 pKa = 4.7KK1125 pKa = 10.87INEE1128 pKa = 4.46CVKK1131 pKa = 10.55SQSFRR1136 pKa = 11.84YY1137 pKa = 8.66GFCGNGTHH1145 pKa = 6.04VFSVVNAAPDD1155 pKa = 3.22GMMFFHH1161 pKa = 7.24SVLLPTAYY1169 pKa = 9.73MEE1171 pKa = 4.19VAAFSGLCVEE1181 pKa = 4.56GNGYY1185 pKa = 7.66VLRR1188 pKa = 11.84DD1189 pKa = 3.41TGNVLFEE1196 pKa = 4.9KK1197 pKa = 10.4NGQYY1201 pKa = 10.86LITARR1206 pKa = 11.84KK1207 pKa = 7.74MFEE1210 pKa = 3.7PRR1212 pKa = 11.84VPQTSDD1218 pKa = 2.95FVQITGCDD1226 pKa = 3.24VVYY1229 pKa = 11.09LNVTRR1234 pKa = 11.84DD1235 pKa = 3.69EE1236 pKa = 4.88LPTVIPDD1243 pKa = 3.98YY1244 pKa = 10.97IDD1246 pKa = 3.46VNSTVEE1252 pKa = 4.81DD1253 pKa = 3.67ILSKK1257 pKa = 10.79LPNRR1261 pKa = 11.84TTPEE1265 pKa = 3.49FDD1267 pKa = 4.23LDD1269 pKa = 3.82IFNATYY1275 pKa = 11.19LNLTGEE1281 pKa = 4.41IADD1284 pKa = 3.56LTARR1288 pKa = 11.84SEE1290 pKa = 4.34SLKK1293 pKa = 9.87NTTLEE1298 pKa = 4.05LKK1300 pKa = 10.58EE1301 pKa = 4.49LIANINATLVDD1312 pKa = 4.66LEE1314 pKa = 4.13WLNRR1318 pKa = 11.84VEE1320 pKa = 5.97TYY1322 pKa = 10.41IKK1324 pKa = 9.23WPWWVWLIIVLVLILFTCLMLFCCCSTGCCGIFSCMASSCGACCDD1369 pKa = 3.2IRR1371 pKa = 11.84GTKK1374 pKa = 8.05LQRR1377 pKa = 11.84YY1378 pKa = 5.97EE1379 pKa = 4.8AIEE1382 pKa = 4.12KK1383 pKa = 10.11VHH1385 pKa = 5.3VQQ1387 pKa = 3.06
Molecular weight: 152.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U1WHF1|A0A0U1WHF1_9ALPC Nucleoprotein OS=BtMr-AlphaCoV/SAX2011 OX=1503289 PE=4 SV=1
MM1 pKa = 7.5SSVKK5 pKa = 10.2FEE7 pKa = 3.95ASGRR11 pKa = 11.84TGRR14 pKa = 11.84TPLSFFAPLTVSSDD28 pKa = 3.05KK29 pKa = 11.16KK30 pKa = 10.3FWNVLPKK37 pKa = 10.7NAVPTGKK44 pKa = 10.51GNAKK48 pKa = 9.27QQVGYY53 pKa = 10.09WNEE56 pKa = 3.72QPRR59 pKa = 11.84WRR61 pKa = 11.84MQRR64 pKa = 11.84GEE66 pKa = 4.18RR67 pKa = 11.84VEE69 pKa = 4.45KK70 pKa = 9.52PSYY73 pKa = 7.31WHH75 pKa = 6.93FYY77 pKa = 10.45FLGTGPHH84 pKa = 6.6ADD86 pKa = 2.73AKK88 pKa = 10.22FRR90 pKa = 11.84EE91 pKa = 4.68RR92 pKa = 11.84IPGVVWVSKK101 pKa = 11.16ANADD105 pKa = 4.36LNPTNLGTRR114 pKa = 11.84SKK116 pKa = 10.24ARR118 pKa = 11.84SVITPKK124 pKa = 10.17FDD126 pKa = 3.67SEE128 pKa = 4.47LPSDD132 pKa = 3.79IEE134 pKa = 4.32IVDD137 pKa = 3.81KK138 pKa = 11.41SSAPNSRR145 pKa = 11.84GNSRR149 pKa = 11.84SQSRR153 pKa = 11.84GPGNGANSRR162 pKa = 11.84NNSKK166 pKa = 10.14GRR168 pKa = 11.84NNTNSRR174 pKa = 11.84DD175 pKa = 3.47NSNNRR180 pKa = 11.84GKK182 pKa = 10.03SQSRR186 pKa = 11.84NSSRR190 pKa = 11.84NRR192 pKa = 11.84GQQRR196 pKa = 11.84NNNQQRR202 pKa = 11.84QGTGAAGNGTADD214 pKa = 3.77LAAAIVLALEE224 pKa = 4.28KK225 pKa = 10.8AGLTRR230 pKa = 11.84EE231 pKa = 4.24TEE233 pKa = 3.98KK234 pKa = 11.17APKK237 pKa = 9.88KK238 pKa = 10.39EE239 pKa = 4.0SANNNKK245 pKa = 9.64KK246 pKa = 10.22SEE248 pKa = 4.14SRR250 pKa = 11.84ASSPAPAQSKK260 pKa = 10.29KK261 pKa = 10.49DD262 pKa = 3.55QLGKK266 pKa = 10.98VMWKK270 pKa = 10.25RR271 pKa = 11.84NPDD274 pKa = 3.12PSFNVTQCFGPRR286 pKa = 11.84SVYY289 pKa = 10.72QNFGDD294 pKa = 3.81EE295 pKa = 4.18DD296 pKa = 4.5AIKK299 pKa = 10.87NGVKK303 pKa = 10.26AKK305 pKa = 10.39HH306 pKa = 5.33FPSWAEE312 pKa = 3.83LTPTPAALLFGSEE325 pKa = 4.5VITVEE330 pKa = 4.59DD331 pKa = 4.18GDD333 pKa = 5.63DD334 pKa = 3.88IVIQYY339 pKa = 9.04NYY341 pKa = 9.45QMRR344 pKa = 11.84VPKK347 pKa = 10.25TMAALQTFLPQVGAYY362 pKa = 10.47ANGDD366 pKa = 3.73NEE368 pKa = 4.72SEE370 pKa = 4.76SGDD373 pKa = 3.63QPGASAAAAAAPEE386 pKa = 4.52PPALPPKK393 pKa = 10.27LNPKK397 pKa = 9.94ADD399 pKa = 3.24AFVPPKK405 pKa = 10.38INPNYY410 pKa = 8.91FDD412 pKa = 3.54GMKK415 pKa = 10.3VEE417 pKa = 4.62IMNKK421 pKa = 9.31TISDD425 pKa = 3.85DD426 pKa = 3.6STAA429 pKa = 3.61
MM1 pKa = 7.5SSVKK5 pKa = 10.2FEE7 pKa = 3.95ASGRR11 pKa = 11.84TGRR14 pKa = 11.84TPLSFFAPLTVSSDD28 pKa = 3.05KK29 pKa = 11.16KK30 pKa = 10.3FWNVLPKK37 pKa = 10.7NAVPTGKK44 pKa = 10.51GNAKK48 pKa = 9.27QQVGYY53 pKa = 10.09WNEE56 pKa = 3.72QPRR59 pKa = 11.84WRR61 pKa = 11.84MQRR64 pKa = 11.84GEE66 pKa = 4.18RR67 pKa = 11.84VEE69 pKa = 4.45KK70 pKa = 9.52PSYY73 pKa = 7.31WHH75 pKa = 6.93FYY77 pKa = 10.45FLGTGPHH84 pKa = 6.6ADD86 pKa = 2.73AKK88 pKa = 10.22FRR90 pKa = 11.84EE91 pKa = 4.68RR92 pKa = 11.84IPGVVWVSKK101 pKa = 11.16ANADD105 pKa = 4.36LNPTNLGTRR114 pKa = 11.84SKK116 pKa = 10.24ARR118 pKa = 11.84SVITPKK124 pKa = 10.17FDD126 pKa = 3.67SEE128 pKa = 4.47LPSDD132 pKa = 3.79IEE134 pKa = 4.32IVDD137 pKa = 3.81KK138 pKa = 11.41SSAPNSRR145 pKa = 11.84GNSRR149 pKa = 11.84SQSRR153 pKa = 11.84GPGNGANSRR162 pKa = 11.84NNSKK166 pKa = 10.14GRR168 pKa = 11.84NNTNSRR174 pKa = 11.84DD175 pKa = 3.47NSNNRR180 pKa = 11.84GKK182 pKa = 10.03SQSRR186 pKa = 11.84NSSRR190 pKa = 11.84NRR192 pKa = 11.84GQQRR196 pKa = 11.84NNNQQRR202 pKa = 11.84QGTGAAGNGTADD214 pKa = 3.77LAAAIVLALEE224 pKa = 4.28KK225 pKa = 10.8AGLTRR230 pKa = 11.84EE231 pKa = 4.24TEE233 pKa = 3.98KK234 pKa = 11.17APKK237 pKa = 9.88KK238 pKa = 10.39EE239 pKa = 4.0SANNNKK245 pKa = 9.64KK246 pKa = 10.22SEE248 pKa = 4.14SRR250 pKa = 11.84ASSPAPAQSKK260 pKa = 10.29KK261 pKa = 10.49DD262 pKa = 3.55QLGKK266 pKa = 10.98VMWKK270 pKa = 10.25RR271 pKa = 11.84NPDD274 pKa = 3.12PSFNVTQCFGPRR286 pKa = 11.84SVYY289 pKa = 10.72QNFGDD294 pKa = 3.81EE295 pKa = 4.18DD296 pKa = 4.5AIKK299 pKa = 10.87NGVKK303 pKa = 10.26AKK305 pKa = 10.39HH306 pKa = 5.33FPSWAEE312 pKa = 3.83LTPTPAALLFGSEE325 pKa = 4.5VITVEE330 pKa = 4.59DD331 pKa = 4.18GDD333 pKa = 5.63DD334 pKa = 3.88IVIQYY339 pKa = 9.04NYY341 pKa = 9.45QMRR344 pKa = 11.84VPKK347 pKa = 10.25TMAALQTFLPQVGAYY362 pKa = 10.47ANGDD366 pKa = 3.73NEE368 pKa = 4.72SEE370 pKa = 4.76SGDD373 pKa = 3.63QPGASAAAAAAPEE386 pKa = 4.52PPALPPKK393 pKa = 10.27LNPKK397 pKa = 9.94ADD399 pKa = 3.24AFVPPKK405 pKa = 10.38INPNYY410 pKa = 8.91FDD412 pKa = 3.54GMKK415 pKa = 10.3VEE417 pKa = 4.62IMNKK421 pKa = 9.31TISDD425 pKa = 3.85DD426 pKa = 3.6STAA429 pKa = 3.61
Molecular weight: 46.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9000 |
76 |
6662 |
1500.0 |
166.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.378 ± 0.482 | 3.478 ± 0.785 |
5.656 ± 0.7 | 3.711 ± 0.364 |
5.511 ± 0.286 | 6.622 ± 0.453 |
1.856 ± 0.279 | 4.767 ± 0.863 |
5.2 ± 0.94 | 8.767 ± 0.829 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.1 ± 0.178 | 5.678 ± 0.532 |
3.589 ± 0.643 | 3.056 ± 0.466 |
3.6 ± 0.351 | 6.956 ± 0.472 |
6.311 ± 0.912 | 9.644 ± 0.602 |
1.3 ± 0.427 | 4.822 ± 0.409 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
