
Sphingobium sp. TKS
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingobium; unclassified Sphingobium
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
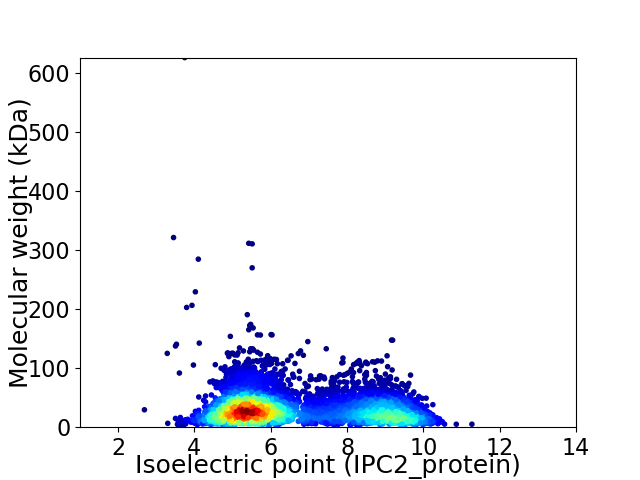
Virtual 2D-PAGE plot for 5795 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126RQQ2|A0A126RQQ2_9SPHN Uncharacterized protein OS=Sphingobium sp. TKS OX=1315974 GN=K426_17410 PE=4 SV=1
MM1 pKa = 7.42SVTMSDD7 pKa = 3.17TALKK11 pKa = 10.48IGDD14 pKa = 3.86TSTVTVAFSEE24 pKa = 4.43AVTGFDD30 pKa = 3.36NSDD33 pKa = 3.2VTVEE37 pKa = 4.32HH38 pKa = 6.49GTLSTFTSLDD48 pKa = 3.43GKK50 pKa = 10.57VWTATFTPTANIEE63 pKa = 4.08DD64 pKa = 4.0SSNVVTVGSGYY75 pKa = 9.32TDD77 pKa = 3.56LAGNAGSGGTSANYY91 pKa = 10.57AIDD94 pKa = 3.66TKK96 pKa = 11.33APTLSITDD104 pKa = 3.83DD105 pKa = 3.37EE106 pKa = 4.66AAAVANNAGGAVVFTFTFSEE126 pKa = 4.4NVTGFTASDD135 pKa = 3.23ITVTGGTAGTFTAVNGHH152 pKa = 6.36SYY154 pKa = 9.33TLSVMPNADD163 pKa = 3.73SQSGTIAVSVADD175 pKa = 4.47GAATDD180 pKa = 3.93LAGNAVTGASASQNYY195 pKa = 7.68DD196 pKa = 3.19TLNPTVTVGFAGTALSDD213 pKa = 3.65GTASSQVNFVFSEE226 pKa = 4.51APVGFAQGDD235 pKa = 3.37ISASHH240 pKa = 6.18GAISNLVMDD249 pKa = 5.76DD250 pKa = 3.21ATHH253 pKa = 5.18YY254 pKa = 10.46HH255 pKa = 5.88ATFTADD261 pKa = 4.09DD262 pKa = 4.52GFSGTGSVTVDD273 pKa = 3.03AARR276 pKa = 11.84FTDD279 pKa = 4.48AALNDD284 pKa = 3.68NAAATPATIDD294 pKa = 3.64IDD296 pKa = 4.14TLNPTLVSVTMSDD309 pKa = 3.22TALKK313 pKa = 10.48IGDD316 pKa = 3.86TSTVTVAFSEE326 pKa = 4.43AVTGFDD332 pKa = 3.36NSDD335 pKa = 3.2VTVEE339 pKa = 4.32HH340 pKa = 6.49GTLSTFTSLDD350 pKa = 3.43GKK352 pKa = 10.57VWTATFTPTANIEE365 pKa = 4.08DD366 pKa = 4.0SSNVVTVGPNYY377 pKa = 10.15TDD379 pKa = 3.57LAGNAGSGGTSANYY393 pKa = 10.57AIDD396 pKa = 3.66TKK398 pKa = 11.33APTLSITDD406 pKa = 3.83DD407 pKa = 3.37EE408 pKa = 4.66AAAVANNAGGAVVFTFTFSEE428 pKa = 4.58DD429 pKa = 3.42VTGFTTDD436 pKa = 4.47DD437 pKa = 3.3ITVTGGTAGTFTAIDD452 pKa = 3.49GHH454 pKa = 6.82SYY456 pKa = 9.22TLSVTPDD463 pKa = 3.23ANNQAGTIAVSVADD477 pKa = 4.12GAAVDD482 pKa = 4.17GAGNPIIGSSASQNYY497 pKa = 7.68DD498 pKa = 3.17TLNPTLASVTMSDD511 pKa = 3.29SALKK515 pKa = 10.54IGDD518 pKa = 3.68TSIVTVTFSEE528 pKa = 4.5AVTGFDD534 pKa = 3.36NSDD537 pKa = 3.2VTVEE541 pKa = 4.32HH542 pKa = 6.49GTLSTFTSLDD552 pKa = 3.43GKK554 pKa = 10.57VWTATFTPTANIEE567 pKa = 4.11DD568 pKa = 4.34DD569 pKa = 4.37SNVVTVGSGYY579 pKa = 9.32TDD581 pKa = 3.57LAGNAGTGGTSANYY595 pKa = 10.19EE596 pKa = 4.22IDD598 pKa = 3.4TKK600 pKa = 11.42APTLSITDD608 pKa = 3.83DD609 pKa = 3.37EE610 pKa = 4.66AAAVANNAGGAVVFTFTFSEE630 pKa = 4.4NVTGFTASDD639 pKa = 3.23ITVTGGTAGTFTAVNGHH656 pKa = 6.41SYY658 pKa = 9.21TLSVTPDD665 pKa = 3.23ANNQAGTIGVSVANGAAMDD684 pKa = 4.17GAGNPIIGASASQNYY699 pKa = 7.48DD700 pKa = 3.1TKK702 pKa = 11.3NPTVAVNIIDD712 pKa = 4.62ASLDD716 pKa = 3.6NNDD719 pKa = 4.4PSSQVTFTFSEE730 pKa = 4.79TPVGFGSGDD739 pKa = 3.0ISAVGGTISNFQSTADD755 pKa = 3.82PLVYY759 pKa = 9.64TATYY763 pKa = 8.41TKK765 pKa = 9.89TAGFTGTGSVSVGSTLYY782 pKa = 10.46TDD784 pKa = 4.4AALNPGAPGSDD795 pKa = 2.99TVAVSSGDD803 pKa = 3.47TTSPGITSITGLQGNNNQNSKK824 pKa = 11.12LIITFNEE831 pKa = 3.66AMNTSFFDD839 pKa = 3.17ASKK842 pKa = 10.31IHH844 pKa = 6.28LAAATGTASIVTGTGAWSSNNTVFTIEE871 pKa = 4.01VAKK874 pKa = 10.47TNGANNGTSYY884 pKa = 9.77TAAIDD889 pKa = 3.35AGAFKK894 pKa = 10.69DD895 pKa = 3.9VAGNLSNAAQASGLKK910 pKa = 9.26PAGVAGAPINLGLSSPADD928 pKa = 3.31QAGDD932 pKa = 3.43VLVTVTHH939 pKa = 6.37MPDD942 pKa = 2.8DD943 pKa = 3.53WALNGGTRR951 pKa = 11.84NADD954 pKa = 3.45GSWTVLTADD963 pKa = 3.82PRR965 pKa = 11.84ALSVTTPSTFAGAMVLDD982 pKa = 4.56VAMSWTQADD991 pKa = 3.56GAMATMHH998 pKa = 6.68IADD1001 pKa = 4.21NVEE1004 pKa = 4.57AYY1006 pKa = 10.52APGNPIFAWSGDD1018 pKa = 3.64DD1019 pKa = 3.44TLTGSAQADD1028 pKa = 3.23IFVFAQPIGADD1039 pKa = 3.33QVHH1042 pKa = 6.98DD1043 pKa = 4.0FDD1045 pKa = 5.45IGADD1049 pKa = 3.81RR1050 pKa = 11.84IDD1052 pKa = 4.58LIGYY1056 pKa = 9.63AGIGDD1061 pKa = 4.18FSGVLAHH1068 pKa = 6.97LAEE1071 pKa = 4.89DD1072 pKa = 3.69ASGNALLTLAEE1083 pKa = 4.54GQTITLQGVTAAALTTANFLFDD1105 pKa = 3.71EE1106 pKa = 4.85TPVVSNSGVMTIGNGAMLPLSGTIVNSGVISLEE1139 pKa = 4.11STGDD1143 pKa = 3.35TTLLQLIQYY1152 pKa = 7.63GTTLQGGGHH1161 pKa = 5.59VVLSDD1166 pKa = 3.61DD1167 pKa = 4.1GGNVISGTLPSVTLNNVDD1185 pKa = 4.44NIISGAGEE1193 pKa = 3.88IGAGTLTLINGGSIIADD1210 pKa = 3.63GTHH1213 pKa = 6.48GLVLDD1218 pKa = 4.47TGTNMISNSGIIAATGSGGLTVVSAIDD1245 pKa = 3.66NEE1247 pKa = 4.4GLIQLNSSHH1256 pKa = 6.5AVLVGDD1262 pKa = 3.82VSGSGDD1268 pKa = 3.51VVLSGTATVEE1278 pKa = 4.17FGGHH1282 pKa = 4.05VTSDD1286 pKa = 2.86IVVDD1290 pKa = 4.59AGATGLILFDD1300 pKa = 5.1DD1301 pKa = 5.11SISFSGTIAGLDD1313 pKa = 3.22SDD1315 pKa = 5.3DD1316 pKa = 5.52RR1317 pKa = 11.84VDD1319 pKa = 5.22LSDD1322 pKa = 4.44LAFDD1326 pKa = 4.16AATMLSYY1333 pKa = 10.8AANDD1337 pKa = 3.59TGTGGVLTVTSGADD1351 pKa = 3.43VAHH1354 pKa = 6.48LTLIGSYY1361 pKa = 9.76HH1362 pKa = 7.39AEE1364 pKa = 4.51DD1365 pKa = 3.76FGLWSDD1371 pKa = 3.79GHH1373 pKa = 6.99SGTLISFGG1381 pKa = 3.65
MM1 pKa = 7.42SVTMSDD7 pKa = 3.17TALKK11 pKa = 10.48IGDD14 pKa = 3.86TSTVTVAFSEE24 pKa = 4.43AVTGFDD30 pKa = 3.36NSDD33 pKa = 3.2VTVEE37 pKa = 4.32HH38 pKa = 6.49GTLSTFTSLDD48 pKa = 3.43GKK50 pKa = 10.57VWTATFTPTANIEE63 pKa = 4.08DD64 pKa = 4.0SSNVVTVGSGYY75 pKa = 9.32TDD77 pKa = 3.56LAGNAGSGGTSANYY91 pKa = 10.57AIDD94 pKa = 3.66TKK96 pKa = 11.33APTLSITDD104 pKa = 3.83DD105 pKa = 3.37EE106 pKa = 4.66AAAVANNAGGAVVFTFTFSEE126 pKa = 4.4NVTGFTASDD135 pKa = 3.23ITVTGGTAGTFTAVNGHH152 pKa = 6.36SYY154 pKa = 9.33TLSVMPNADD163 pKa = 3.73SQSGTIAVSVADD175 pKa = 4.47GAATDD180 pKa = 3.93LAGNAVTGASASQNYY195 pKa = 7.68DD196 pKa = 3.19TLNPTVTVGFAGTALSDD213 pKa = 3.65GTASSQVNFVFSEE226 pKa = 4.51APVGFAQGDD235 pKa = 3.37ISASHH240 pKa = 6.18GAISNLVMDD249 pKa = 5.76DD250 pKa = 3.21ATHH253 pKa = 5.18YY254 pKa = 10.46HH255 pKa = 5.88ATFTADD261 pKa = 4.09DD262 pKa = 4.52GFSGTGSVTVDD273 pKa = 3.03AARR276 pKa = 11.84FTDD279 pKa = 4.48AALNDD284 pKa = 3.68NAAATPATIDD294 pKa = 3.64IDD296 pKa = 4.14TLNPTLVSVTMSDD309 pKa = 3.22TALKK313 pKa = 10.48IGDD316 pKa = 3.86TSTVTVAFSEE326 pKa = 4.43AVTGFDD332 pKa = 3.36NSDD335 pKa = 3.2VTVEE339 pKa = 4.32HH340 pKa = 6.49GTLSTFTSLDD350 pKa = 3.43GKK352 pKa = 10.57VWTATFTPTANIEE365 pKa = 4.08DD366 pKa = 4.0SSNVVTVGPNYY377 pKa = 10.15TDD379 pKa = 3.57LAGNAGSGGTSANYY393 pKa = 10.57AIDD396 pKa = 3.66TKK398 pKa = 11.33APTLSITDD406 pKa = 3.83DD407 pKa = 3.37EE408 pKa = 4.66AAAVANNAGGAVVFTFTFSEE428 pKa = 4.58DD429 pKa = 3.42VTGFTTDD436 pKa = 4.47DD437 pKa = 3.3ITVTGGTAGTFTAIDD452 pKa = 3.49GHH454 pKa = 6.82SYY456 pKa = 9.22TLSVTPDD463 pKa = 3.23ANNQAGTIAVSVADD477 pKa = 4.12GAAVDD482 pKa = 4.17GAGNPIIGSSASQNYY497 pKa = 7.68DD498 pKa = 3.17TLNPTLASVTMSDD511 pKa = 3.29SALKK515 pKa = 10.54IGDD518 pKa = 3.68TSIVTVTFSEE528 pKa = 4.5AVTGFDD534 pKa = 3.36NSDD537 pKa = 3.2VTVEE541 pKa = 4.32HH542 pKa = 6.49GTLSTFTSLDD552 pKa = 3.43GKK554 pKa = 10.57VWTATFTPTANIEE567 pKa = 4.11DD568 pKa = 4.34DD569 pKa = 4.37SNVVTVGSGYY579 pKa = 9.32TDD581 pKa = 3.57LAGNAGTGGTSANYY595 pKa = 10.19EE596 pKa = 4.22IDD598 pKa = 3.4TKK600 pKa = 11.42APTLSITDD608 pKa = 3.83DD609 pKa = 3.37EE610 pKa = 4.66AAAVANNAGGAVVFTFTFSEE630 pKa = 4.4NVTGFTASDD639 pKa = 3.23ITVTGGTAGTFTAVNGHH656 pKa = 6.41SYY658 pKa = 9.21TLSVTPDD665 pKa = 3.23ANNQAGTIGVSVANGAAMDD684 pKa = 4.17GAGNPIIGASASQNYY699 pKa = 7.48DD700 pKa = 3.1TKK702 pKa = 11.3NPTVAVNIIDD712 pKa = 4.62ASLDD716 pKa = 3.6NNDD719 pKa = 4.4PSSQVTFTFSEE730 pKa = 4.79TPVGFGSGDD739 pKa = 3.0ISAVGGTISNFQSTADD755 pKa = 3.82PLVYY759 pKa = 9.64TATYY763 pKa = 8.41TKK765 pKa = 9.89TAGFTGTGSVSVGSTLYY782 pKa = 10.46TDD784 pKa = 4.4AALNPGAPGSDD795 pKa = 2.99TVAVSSGDD803 pKa = 3.47TTSPGITSITGLQGNNNQNSKK824 pKa = 11.12LIITFNEE831 pKa = 3.66AMNTSFFDD839 pKa = 3.17ASKK842 pKa = 10.31IHH844 pKa = 6.28LAAATGTASIVTGTGAWSSNNTVFTIEE871 pKa = 4.01VAKK874 pKa = 10.47TNGANNGTSYY884 pKa = 9.77TAAIDD889 pKa = 3.35AGAFKK894 pKa = 10.69DD895 pKa = 3.9VAGNLSNAAQASGLKK910 pKa = 9.26PAGVAGAPINLGLSSPADD928 pKa = 3.31QAGDD932 pKa = 3.43VLVTVTHH939 pKa = 6.37MPDD942 pKa = 2.8DD943 pKa = 3.53WALNGGTRR951 pKa = 11.84NADD954 pKa = 3.45GSWTVLTADD963 pKa = 3.82PRR965 pKa = 11.84ALSVTTPSTFAGAMVLDD982 pKa = 4.56VAMSWTQADD991 pKa = 3.56GAMATMHH998 pKa = 6.68IADD1001 pKa = 4.21NVEE1004 pKa = 4.57AYY1006 pKa = 10.52APGNPIFAWSGDD1018 pKa = 3.64DD1019 pKa = 3.44TLTGSAQADD1028 pKa = 3.23IFVFAQPIGADD1039 pKa = 3.33QVHH1042 pKa = 6.98DD1043 pKa = 4.0FDD1045 pKa = 5.45IGADD1049 pKa = 3.81RR1050 pKa = 11.84IDD1052 pKa = 4.58LIGYY1056 pKa = 9.63AGIGDD1061 pKa = 4.18FSGVLAHH1068 pKa = 6.97LAEE1071 pKa = 4.89DD1072 pKa = 3.69ASGNALLTLAEE1083 pKa = 4.54GQTITLQGVTAAALTTANFLFDD1105 pKa = 3.71EE1106 pKa = 4.85TPVVSNSGVMTIGNGAMLPLSGTIVNSGVISLEE1139 pKa = 4.11STGDD1143 pKa = 3.35TTLLQLIQYY1152 pKa = 7.63GTTLQGGGHH1161 pKa = 5.59VVLSDD1166 pKa = 3.61DD1167 pKa = 4.1GGNVISGTLPSVTLNNVDD1185 pKa = 4.44NIISGAGEE1193 pKa = 3.88IGAGTLTLINGGSIIADD1210 pKa = 3.63GTHH1213 pKa = 6.48GLVLDD1218 pKa = 4.47TGTNMISNSGIIAATGSGGLTVVSAIDD1245 pKa = 3.66NEE1247 pKa = 4.4GLIQLNSSHH1256 pKa = 6.5AVLVGDD1262 pKa = 3.82VSGSGDD1268 pKa = 3.51VVLSGTATVEE1278 pKa = 4.17FGGHH1282 pKa = 4.05VTSDD1286 pKa = 2.86IVVDD1290 pKa = 4.59AGATGLILFDD1300 pKa = 5.1DD1301 pKa = 5.11SISFSGTIAGLDD1313 pKa = 3.22SDD1315 pKa = 5.3DD1316 pKa = 5.52RR1317 pKa = 11.84VDD1319 pKa = 5.22LSDD1322 pKa = 4.44LAFDD1326 pKa = 4.16AATMLSYY1333 pKa = 10.8AANDD1337 pKa = 3.59TGTGGVLTVTSGADD1351 pKa = 3.43VAHH1354 pKa = 6.48LTLIGSYY1361 pKa = 9.76HH1362 pKa = 7.39AEE1364 pKa = 4.51DD1365 pKa = 3.76FGLWSDD1371 pKa = 3.79GHH1373 pKa = 6.99SGTLISFGG1381 pKa = 3.65
Molecular weight: 137.54 kDa
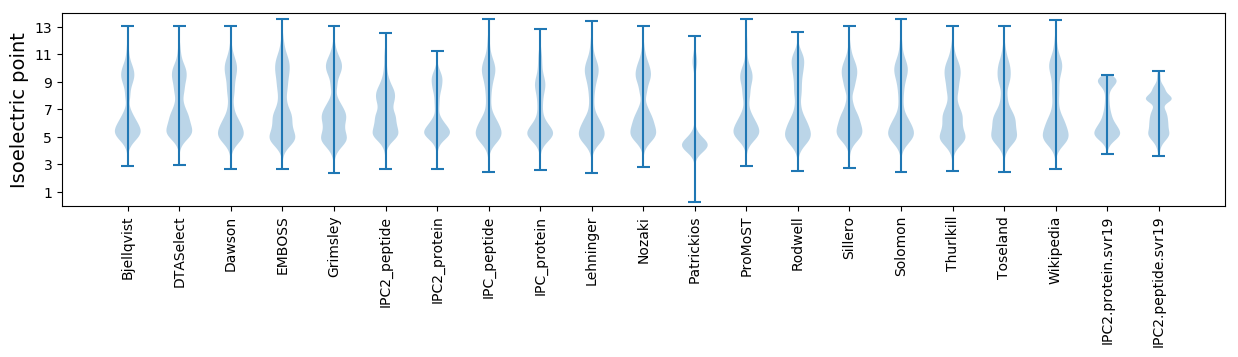
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126RQT2|A0A126RQT2_9SPHN HTH_17 domain-containing protein OS=Sphingobium sp. TKS OX=1315974 GN=K426_18505 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATPGGRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.06SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATPGGRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.06SLSAA44 pKa = 3.93
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1774848 |
23 |
7080 |
306.3 |
33.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.908 ± 0.046 | 0.848 ± 0.011 |
5.941 ± 0.026 | 5.466 ± 0.035 |
3.494 ± 0.02 | 8.794 ± 0.103 |
2.138 ± 0.021 | 5.182 ± 0.021 |
3.053 ± 0.025 | 10.024 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.52 ± 0.018 | 2.539 ± 0.022 |
5.189 ± 0.032 | 3.263 ± 0.019 |
7.511 ± 0.047 | 5.491 ± 0.032 |
5.095 ± 0.031 | 6.919 ± 0.03 |
1.415 ± 0.015 | 2.21 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
