
Bornean orang-utan polyomavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Alphapolyomavirus; Pongo pygmaeus polyomavirus 1
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
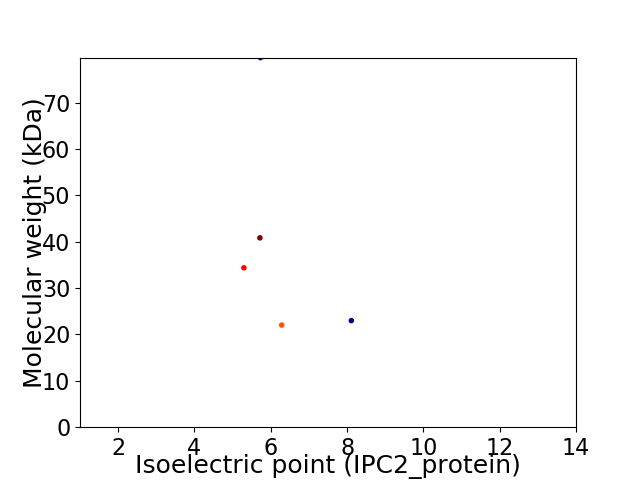
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
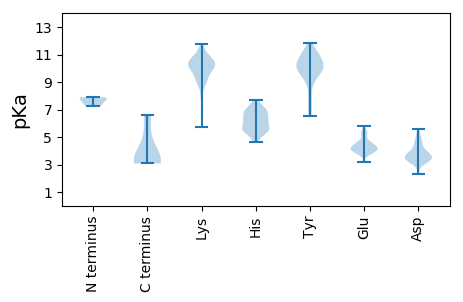
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C9X3W4|C9X3W4_9POLY Minor capsid protein VP2 OS=Bornean orang-utan polyomavirus OX=1604874 GN=VP3 PE=3 SV=1
MM1 pKa = 7.47GGVISLFFDD10 pKa = 3.03IVEE13 pKa = 4.39ISAEE17 pKa = 4.01LSTATGFAVDD27 pKa = 3.86AVLTGEE33 pKa = 3.94AAAAVEE39 pKa = 4.36TEE41 pKa = 4.29VMGLMTIEE49 pKa = 4.43GLSASEE55 pKa = 4.18ALGAMGLTMEE65 pKa = 5.37DD66 pKa = 3.84FSLMHH71 pKa = 7.26ALPGMLSEE79 pKa = 4.54AVGIGTLFQAISGASGLVAAGIRR102 pKa = 11.84YY103 pKa = 9.05GYY105 pKa = 10.05ARR107 pKa = 11.84EE108 pKa = 3.77VSIVNRR114 pKa = 11.84NKK116 pKa = 10.49RR117 pKa = 11.84NPSEE121 pKa = 3.93MALQVWRR128 pKa = 11.84PWDD131 pKa = 3.54YY132 pKa = 11.83YY133 pKa = 11.28DD134 pKa = 4.6ILFPGVNTFAHH145 pKa = 6.17YY146 pKa = 11.16LNILDD151 pKa = 4.79DD152 pKa = 3.9WASSLIHH159 pKa = 5.36TVSRR163 pKa = 11.84YY164 pKa = 7.19VWDD167 pKa = 5.59AILHH171 pKa = 5.7EE172 pKa = 4.69SRR174 pKa = 11.84HH175 pKa = 5.42QIGHH179 pKa = 7.09ASRR182 pKa = 11.84EE183 pKa = 4.1LVARR187 pKa = 11.84GSGHH191 pKa = 6.03FQDD194 pKa = 5.13LMARR198 pKa = 11.84LIEE201 pKa = 3.94NSRR204 pKa = 11.84WVLTTGPSNVYY215 pKa = 10.12SHH217 pKa = 7.1LEE219 pKa = 3.67SYY221 pKa = 10.31YY222 pKa = 10.22RR223 pKa = 11.84QLPGISPPQARR234 pKa = 11.84DD235 pKa = 3.15LYY237 pKa = 11.02RR238 pKa = 11.84RR239 pKa = 11.84LQEE242 pKa = 4.67KK243 pKa = 10.35IPDD246 pKa = 3.54RR247 pKa = 11.84HH248 pKa = 5.44QLAVATDD255 pKa = 3.81EE256 pKa = 4.14SAEE259 pKa = 4.18VIEE262 pKa = 5.07TYY264 pKa = 9.25PAPGGAHH271 pKa = 5.75QRR273 pKa = 11.84VCPDD277 pKa = 2.28WMLPLVLGLYY287 pKa = 10.89GDD289 pKa = 3.78ITPTFGEE296 pKa = 4.38YY297 pKa = 9.83LRR299 pKa = 11.84KK300 pKa = 9.72EE301 pKa = 4.37DD302 pKa = 3.8GPQKK306 pKa = 10.2KK307 pKa = 9.18RR308 pKa = 11.84RR309 pKa = 11.84RR310 pKa = 11.84LL311 pKa = 3.33
MM1 pKa = 7.47GGVISLFFDD10 pKa = 3.03IVEE13 pKa = 4.39ISAEE17 pKa = 4.01LSTATGFAVDD27 pKa = 3.86AVLTGEE33 pKa = 3.94AAAAVEE39 pKa = 4.36TEE41 pKa = 4.29VMGLMTIEE49 pKa = 4.43GLSASEE55 pKa = 4.18ALGAMGLTMEE65 pKa = 5.37DD66 pKa = 3.84FSLMHH71 pKa = 7.26ALPGMLSEE79 pKa = 4.54AVGIGTLFQAISGASGLVAAGIRR102 pKa = 11.84YY103 pKa = 9.05GYY105 pKa = 10.05ARR107 pKa = 11.84EE108 pKa = 3.77VSIVNRR114 pKa = 11.84NKK116 pKa = 10.49RR117 pKa = 11.84NPSEE121 pKa = 3.93MALQVWRR128 pKa = 11.84PWDD131 pKa = 3.54YY132 pKa = 11.83YY133 pKa = 11.28DD134 pKa = 4.6ILFPGVNTFAHH145 pKa = 6.17YY146 pKa = 11.16LNILDD151 pKa = 4.79DD152 pKa = 3.9WASSLIHH159 pKa = 5.36TVSRR163 pKa = 11.84YY164 pKa = 7.19VWDD167 pKa = 5.59AILHH171 pKa = 5.7EE172 pKa = 4.69SRR174 pKa = 11.84HH175 pKa = 5.42QIGHH179 pKa = 7.09ASRR182 pKa = 11.84EE183 pKa = 4.1LVARR187 pKa = 11.84GSGHH191 pKa = 6.03FQDD194 pKa = 5.13LMARR198 pKa = 11.84LIEE201 pKa = 3.94NSRR204 pKa = 11.84WVLTTGPSNVYY215 pKa = 10.12SHH217 pKa = 7.1LEE219 pKa = 3.67SYY221 pKa = 10.31YY222 pKa = 10.22RR223 pKa = 11.84QLPGISPPQARR234 pKa = 11.84DD235 pKa = 3.15LYY237 pKa = 11.02RR238 pKa = 11.84RR239 pKa = 11.84LQEE242 pKa = 4.67KK243 pKa = 10.35IPDD246 pKa = 3.54RR247 pKa = 11.84HH248 pKa = 5.44QLAVATDD255 pKa = 3.81EE256 pKa = 4.14SAEE259 pKa = 4.18VIEE262 pKa = 5.07TYY264 pKa = 9.25PAPGGAHH271 pKa = 5.75QRR273 pKa = 11.84VCPDD277 pKa = 2.28WMLPLVLGLYY287 pKa = 10.89GDD289 pKa = 3.78ITPTFGEE296 pKa = 4.38YY297 pKa = 9.83LRR299 pKa = 11.84KK300 pKa = 9.72EE301 pKa = 4.37DD302 pKa = 3.8GPQKK306 pKa = 10.2KK307 pKa = 9.18RR308 pKa = 11.84RR309 pKa = 11.84RR310 pKa = 11.84LL311 pKa = 3.33
Molecular weight: 34.38 kDa
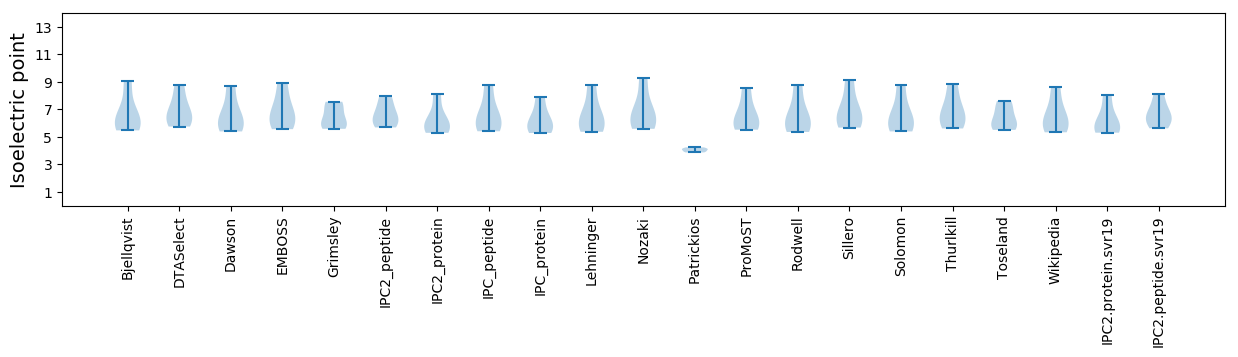
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C9X3W7|C9X3W7_9POLY Small T antigen OS=Bornean orang-utan polyomavirus OX=1604874 GN=small T PE=4 SV=1
MM1 pKa = 7.9DD2 pKa = 5.14RR3 pKa = 11.84FLSRR7 pKa = 11.84EE8 pKa = 3.77EE9 pKa = 4.32SNEE12 pKa = 3.71LMEE15 pKa = 5.31LLNIPKK21 pKa = 9.59HH22 pKa = 6.19CYY24 pKa = 10.64GNFALMKK31 pKa = 9.1INHH34 pKa = 6.7RR35 pKa = 11.84KK36 pKa = 8.83MSLKK40 pKa = 9.85YY41 pKa = 10.36HH42 pKa = 6.84PDD44 pKa = 3.2KK45 pKa = 11.6GGDD48 pKa = 3.64PEE50 pKa = 5.33KK51 pKa = 10.19MARR54 pKa = 11.84LNALWQKK61 pKa = 10.07LQEE64 pKa = 4.64GIYY67 pKa = 9.98NARR70 pKa = 11.84QEE72 pKa = 4.4YY73 pKa = 6.51PTSFGSKK80 pKa = 7.83VGSWFWEE87 pKa = 4.03ANLITLKK94 pKa = 10.47EE95 pKa = 4.17YY96 pKa = 9.7YY97 pKa = 10.08GKK99 pKa = 10.64RR100 pKa = 11.84KK101 pKa = 9.54FDD103 pKa = 3.69EE104 pKa = 4.2NLIKK108 pKa = 10.33HH109 pKa = 6.16WPQCAEE115 pKa = 4.47KK116 pKa = 10.54ALQEE120 pKa = 4.59CKK122 pKa = 10.67CLTCKK127 pKa = 10.55LGLQHH132 pKa = 6.62QVYY135 pKa = 9.86KK136 pKa = 10.87QMHH139 pKa = 4.88QKK141 pKa = 10.21KK142 pKa = 9.29CIVWGEE148 pKa = 4.14CFCFKK153 pKa = 10.49CYY155 pKa = 10.45CIWFGRR161 pKa = 11.84GPLLSGFLVGMEE173 pKa = 4.51CNCGRR178 pKa = 11.84SGLSSGEE185 pKa = 4.23HH186 pKa = 5.44ISEE189 pKa = 4.05NKK191 pKa = 9.16PRR193 pKa = 11.84PKK195 pKa = 10.49LGG197 pKa = 3.3
MM1 pKa = 7.9DD2 pKa = 5.14RR3 pKa = 11.84FLSRR7 pKa = 11.84EE8 pKa = 3.77EE9 pKa = 4.32SNEE12 pKa = 3.71LMEE15 pKa = 5.31LLNIPKK21 pKa = 9.59HH22 pKa = 6.19CYY24 pKa = 10.64GNFALMKK31 pKa = 9.1INHH34 pKa = 6.7RR35 pKa = 11.84KK36 pKa = 8.83MSLKK40 pKa = 9.85YY41 pKa = 10.36HH42 pKa = 6.84PDD44 pKa = 3.2KK45 pKa = 11.6GGDD48 pKa = 3.64PEE50 pKa = 5.33KK51 pKa = 10.19MARR54 pKa = 11.84LNALWQKK61 pKa = 10.07LQEE64 pKa = 4.64GIYY67 pKa = 9.98NARR70 pKa = 11.84QEE72 pKa = 4.4YY73 pKa = 6.51PTSFGSKK80 pKa = 7.83VGSWFWEE87 pKa = 4.03ANLITLKK94 pKa = 10.47EE95 pKa = 4.17YY96 pKa = 9.7YY97 pKa = 10.08GKK99 pKa = 10.64RR100 pKa = 11.84KK101 pKa = 9.54FDD103 pKa = 3.69EE104 pKa = 4.2NLIKK108 pKa = 10.33HH109 pKa = 6.16WPQCAEE115 pKa = 4.47KK116 pKa = 10.54ALQEE120 pKa = 4.59CKK122 pKa = 10.67CLTCKK127 pKa = 10.55LGLQHH132 pKa = 6.62QVYY135 pKa = 9.86KK136 pKa = 10.87QMHH139 pKa = 4.88QKK141 pKa = 10.21KK142 pKa = 9.29CIVWGEE148 pKa = 4.14CFCFKK153 pKa = 10.49CYY155 pKa = 10.45CIWFGRR161 pKa = 11.84GPLLSGFLVGMEE173 pKa = 4.51CNCGRR178 pKa = 11.84SGLSSGEE185 pKa = 4.23HH186 pKa = 5.44ISEE189 pKa = 4.05NKK191 pKa = 9.16PRR193 pKa = 11.84PKK195 pKa = 10.49LGG197 pKa = 3.3
Molecular weight: 22.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1757 |
190 |
693 |
351.4 |
39.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.464 ± 1.141 | 2.447 ± 0.692 |
5.236 ± 0.543 | 7.057 ± 0.706 |
4.269 ± 0.713 | 6.659 ± 1.195 |
2.675 ± 0.437 | 4.326 ± 0.412 |
6.261 ± 1.375 | 10.074 ± 0.764 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.903 ± 0.468 | 4.496 ± 0.74 |
6.033 ± 0.686 | 3.813 ± 0.226 |
5.236 ± 0.652 | 6.261 ± 0.353 |
5.065 ± 0.697 | 5.976 ± 0.741 |
1.651 ± 0.362 | 4.098 ± 0.436 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
