
Rhizoctonia solani dsRNA virus 3
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Alphapartitivirus; unclassified Alphapartitivirus
Average proteome isoelectric point is 7.1
Get precalculated fractions of proteins
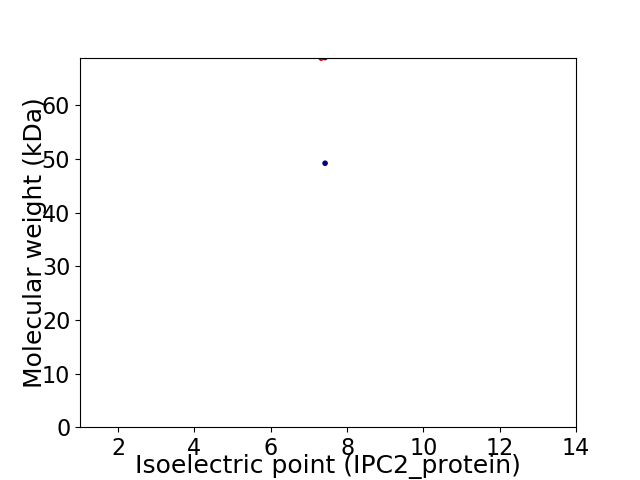
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
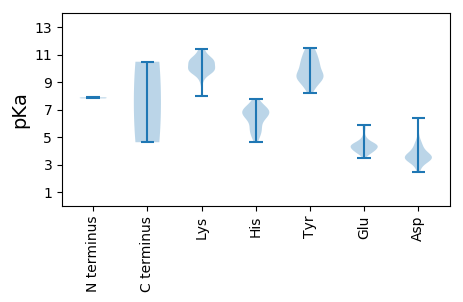
Protein with the lowest isoelectric point:
>tr|A0A143BRM9|A0A143BRM9_9VIRU Coat protein OS=Rhizoctonia solani dsRNA virus 3 OX=1825688 PE=4 SV=1
MM1 pKa = 7.81FYY3 pKa = 8.23TTISTIKK10 pKa = 10.51VFLKK14 pKa = 10.71NLLFSDD20 pKa = 3.25PHH22 pKa = 7.74KK23 pKa = 10.56FVNNFQFIGYY33 pKa = 9.11ASDD36 pKa = 4.38RR37 pKa = 11.84IRR39 pKa = 11.84VSIPYY44 pKa = 9.31RR45 pKa = 11.84DD46 pKa = 3.27EE47 pKa = 4.07FQYY50 pKa = 11.08EE51 pKa = 4.34RR52 pKa = 11.84YY53 pKa = 9.44QRR55 pKa = 11.84TVRR58 pKa = 11.84HH59 pKa = 5.55ALRR62 pKa = 11.84RR63 pKa = 11.84NLIGYY68 pKa = 8.77DD69 pKa = 2.96AEE71 pKa = 4.39YY72 pKa = 10.06IIKK75 pKa = 9.88EE76 pKa = 3.92FHH78 pKa = 6.91HH79 pKa = 6.65PVANLDD85 pKa = 3.73FMVDD89 pKa = 3.45ALRR92 pKa = 11.84KK93 pKa = 9.86GDD95 pKa = 4.42LPDD98 pKa = 3.53HH99 pKa = 6.66VIPKK103 pKa = 9.82DD104 pKa = 3.45EE105 pKa = 4.91HH106 pKa = 6.25YY107 pKa = 11.44SKK109 pKa = 11.38AFAQAAEE116 pKa = 4.41MFRR119 pKa = 11.84PPQLVRR125 pKa = 11.84PVHH128 pKa = 5.75FADD131 pKa = 3.7LRR133 pKa = 11.84MYY135 pKa = 9.65KK136 pKa = 9.8WNWHH140 pKa = 6.25PNVEE144 pKa = 4.19EE145 pKa = 4.41PFYY148 pKa = 11.41SDD150 pKa = 3.76ADD152 pKa = 4.45LIRR155 pKa = 11.84AVSMAAEE162 pKa = 4.65AGLLPDD168 pKa = 4.08ARR170 pKa = 11.84MSFGNLRR177 pKa = 11.84NVVFIKK183 pKa = 10.72ARR185 pKa = 11.84LFLHH189 pKa = 5.67QIKK192 pKa = 10.19RR193 pKa = 11.84KK194 pKa = 9.95QITNPATLWPMMKK207 pKa = 9.27IHH209 pKa = 6.67VKK211 pKa = 9.79PALTKK216 pKa = 9.81VDD218 pKa = 3.63EE219 pKa = 4.5TKK221 pKa = 10.66VRR223 pKa = 11.84IIYY226 pKa = 9.35GVSKK230 pKa = 9.92LHH232 pKa = 6.64VMAQAMFLWPLFNYY246 pKa = 9.38YY247 pKa = 9.84INSDD251 pKa = 4.49DD252 pKa = 6.37DD253 pKa = 3.99PLLWGFEE260 pKa = 4.31TILGGMQKK268 pKa = 9.88LHH270 pKa = 7.13NIMSIPRR277 pKa = 11.84LYY279 pKa = 10.17FQTFVTVDD287 pKa = 2.64WSGFDD292 pKa = 3.49LRR294 pKa = 11.84SVFSLQRR301 pKa = 11.84EE302 pKa = 4.36VFDD305 pKa = 3.01VWRR308 pKa = 11.84TYY310 pKa = 11.16FDD312 pKa = 4.11FNNGYY317 pKa = 9.67IPTKK321 pKa = 10.36FYY323 pKa = 9.14RR324 pKa = 11.84TSVADD329 pKa = 3.97PDD331 pKa = 3.83HH332 pKa = 7.41LEE334 pKa = 5.5ALWEE338 pKa = 4.14WQRR341 pKa = 11.84EE342 pKa = 4.43ACFKK346 pKa = 10.54MPFVMPDD353 pKa = 2.46RR354 pKa = 11.84TMYY357 pKa = 10.72NRR359 pKa = 11.84LFRR362 pKa = 11.84CIPSGLFSTQFLDD375 pKa = 3.51SHH377 pKa = 6.93VNLVMILTILDD388 pKa = 3.56AMHH391 pKa = 6.96FDD393 pKa = 3.03ISKK396 pKa = 10.2IKK398 pKa = 10.3IYY400 pKa = 10.73VQGDD404 pKa = 3.46DD405 pKa = 4.77SIVMLIFHH413 pKa = 7.06IPADD417 pKa = 3.41QHH419 pKa = 7.02IKK421 pKa = 10.34FKK423 pKa = 11.31SDD425 pKa = 3.36FEE427 pKa = 4.47VLAKK431 pKa = 10.76YY432 pKa = 10.69YY433 pKa = 9.45FDD435 pKa = 3.64HH436 pKa = 6.55VARR439 pKa = 11.84PEE441 pKa = 3.97KK442 pKa = 9.78TDD444 pKa = 3.36VYY446 pKa = 9.02EE447 pKa = 4.34TPQGVEE453 pKa = 3.51VLGYY457 pKa = 10.28RR458 pKa = 11.84NYY460 pKa = 10.93NGYY463 pKa = 8.78PEE465 pKa = 4.74RR466 pKa = 11.84DD467 pKa = 3.11WRR469 pKa = 11.84KK470 pKa = 9.62LLAQLLHH477 pKa = 6.34PRR479 pKa = 11.84GALSLEE485 pKa = 4.03TLAARR490 pKa = 11.84CCGIAYY496 pKa = 10.26ASMYY500 pKa = 10.46RR501 pKa = 11.84NPEE504 pKa = 4.06VINVCKK510 pKa = 10.51DD511 pKa = 2.59IYY513 pKa = 11.35NYY515 pKa = 9.24LTTKK519 pKa = 10.44RR520 pKa = 11.84NVVPGEE526 pKa = 3.78LRR528 pKa = 11.84AQRR531 pKa = 11.84DD532 pKa = 3.42IILFGEE538 pKa = 4.58HH539 pKa = 6.49EE540 pKa = 4.38FSIPTDD546 pKa = 3.42HH547 pKa = 7.1FPEE550 pKa = 5.88RR551 pKa = 11.84DD552 pKa = 3.13EE553 pKa = 4.26VTRR556 pKa = 11.84HH557 pKa = 4.99LRR559 pKa = 11.84IPYY562 pKa = 9.45VRR564 pKa = 11.84TDD566 pKa = 3.04SDD568 pKa = 4.24KK569 pKa = 11.34NDD571 pKa = 3.49YY572 pKa = 9.13WPSGHH577 pKa = 6.64FLSLYY582 pKa = 10.48
MM1 pKa = 7.81FYY3 pKa = 8.23TTISTIKK10 pKa = 10.51VFLKK14 pKa = 10.71NLLFSDD20 pKa = 3.25PHH22 pKa = 7.74KK23 pKa = 10.56FVNNFQFIGYY33 pKa = 9.11ASDD36 pKa = 4.38RR37 pKa = 11.84IRR39 pKa = 11.84VSIPYY44 pKa = 9.31RR45 pKa = 11.84DD46 pKa = 3.27EE47 pKa = 4.07FQYY50 pKa = 11.08EE51 pKa = 4.34RR52 pKa = 11.84YY53 pKa = 9.44QRR55 pKa = 11.84TVRR58 pKa = 11.84HH59 pKa = 5.55ALRR62 pKa = 11.84RR63 pKa = 11.84NLIGYY68 pKa = 8.77DD69 pKa = 2.96AEE71 pKa = 4.39YY72 pKa = 10.06IIKK75 pKa = 9.88EE76 pKa = 3.92FHH78 pKa = 6.91HH79 pKa = 6.65PVANLDD85 pKa = 3.73FMVDD89 pKa = 3.45ALRR92 pKa = 11.84KK93 pKa = 9.86GDD95 pKa = 4.42LPDD98 pKa = 3.53HH99 pKa = 6.66VIPKK103 pKa = 9.82DD104 pKa = 3.45EE105 pKa = 4.91HH106 pKa = 6.25YY107 pKa = 11.44SKK109 pKa = 11.38AFAQAAEE116 pKa = 4.41MFRR119 pKa = 11.84PPQLVRR125 pKa = 11.84PVHH128 pKa = 5.75FADD131 pKa = 3.7LRR133 pKa = 11.84MYY135 pKa = 9.65KK136 pKa = 9.8WNWHH140 pKa = 6.25PNVEE144 pKa = 4.19EE145 pKa = 4.41PFYY148 pKa = 11.41SDD150 pKa = 3.76ADD152 pKa = 4.45LIRR155 pKa = 11.84AVSMAAEE162 pKa = 4.65AGLLPDD168 pKa = 4.08ARR170 pKa = 11.84MSFGNLRR177 pKa = 11.84NVVFIKK183 pKa = 10.72ARR185 pKa = 11.84LFLHH189 pKa = 5.67QIKK192 pKa = 10.19RR193 pKa = 11.84KK194 pKa = 9.95QITNPATLWPMMKK207 pKa = 9.27IHH209 pKa = 6.67VKK211 pKa = 9.79PALTKK216 pKa = 9.81VDD218 pKa = 3.63EE219 pKa = 4.5TKK221 pKa = 10.66VRR223 pKa = 11.84IIYY226 pKa = 9.35GVSKK230 pKa = 9.92LHH232 pKa = 6.64VMAQAMFLWPLFNYY246 pKa = 9.38YY247 pKa = 9.84INSDD251 pKa = 4.49DD252 pKa = 6.37DD253 pKa = 3.99PLLWGFEE260 pKa = 4.31TILGGMQKK268 pKa = 9.88LHH270 pKa = 7.13NIMSIPRR277 pKa = 11.84LYY279 pKa = 10.17FQTFVTVDD287 pKa = 2.64WSGFDD292 pKa = 3.49LRR294 pKa = 11.84SVFSLQRR301 pKa = 11.84EE302 pKa = 4.36VFDD305 pKa = 3.01VWRR308 pKa = 11.84TYY310 pKa = 11.16FDD312 pKa = 4.11FNNGYY317 pKa = 9.67IPTKK321 pKa = 10.36FYY323 pKa = 9.14RR324 pKa = 11.84TSVADD329 pKa = 3.97PDD331 pKa = 3.83HH332 pKa = 7.41LEE334 pKa = 5.5ALWEE338 pKa = 4.14WQRR341 pKa = 11.84EE342 pKa = 4.43ACFKK346 pKa = 10.54MPFVMPDD353 pKa = 2.46RR354 pKa = 11.84TMYY357 pKa = 10.72NRR359 pKa = 11.84LFRR362 pKa = 11.84CIPSGLFSTQFLDD375 pKa = 3.51SHH377 pKa = 6.93VNLVMILTILDD388 pKa = 3.56AMHH391 pKa = 6.96FDD393 pKa = 3.03ISKK396 pKa = 10.2IKK398 pKa = 10.3IYY400 pKa = 10.73VQGDD404 pKa = 3.46DD405 pKa = 4.77SIVMLIFHH413 pKa = 7.06IPADD417 pKa = 3.41QHH419 pKa = 7.02IKK421 pKa = 10.34FKK423 pKa = 11.31SDD425 pKa = 3.36FEE427 pKa = 4.47VLAKK431 pKa = 10.76YY432 pKa = 10.69YY433 pKa = 9.45FDD435 pKa = 3.64HH436 pKa = 6.55VARR439 pKa = 11.84PEE441 pKa = 3.97KK442 pKa = 9.78TDD444 pKa = 3.36VYY446 pKa = 9.02EE447 pKa = 4.34TPQGVEE453 pKa = 3.51VLGYY457 pKa = 10.28RR458 pKa = 11.84NYY460 pKa = 10.93NGYY463 pKa = 8.78PEE465 pKa = 4.74RR466 pKa = 11.84DD467 pKa = 3.11WRR469 pKa = 11.84KK470 pKa = 9.62LLAQLLHH477 pKa = 6.34PRR479 pKa = 11.84GALSLEE485 pKa = 4.03TLAARR490 pKa = 11.84CCGIAYY496 pKa = 10.26ASMYY500 pKa = 10.46RR501 pKa = 11.84NPEE504 pKa = 4.06VINVCKK510 pKa = 10.51DD511 pKa = 2.59IYY513 pKa = 11.35NYY515 pKa = 9.24LTTKK519 pKa = 10.44RR520 pKa = 11.84NVVPGEE526 pKa = 3.78LRR528 pKa = 11.84AQRR531 pKa = 11.84DD532 pKa = 3.42IILFGEE538 pKa = 4.58HH539 pKa = 6.49EE540 pKa = 4.38FSIPTDD546 pKa = 3.42HH547 pKa = 7.1FPEE550 pKa = 5.88RR551 pKa = 11.84DD552 pKa = 3.13EE553 pKa = 4.26VTRR556 pKa = 11.84HH557 pKa = 4.99LRR559 pKa = 11.84IPYY562 pKa = 9.45VRR564 pKa = 11.84TDD566 pKa = 3.04SDD568 pKa = 4.24KK569 pKa = 11.34NDD571 pKa = 3.49YY572 pKa = 9.13WPSGHH577 pKa = 6.64FLSLYY582 pKa = 10.48
Molecular weight: 68.77 kDa
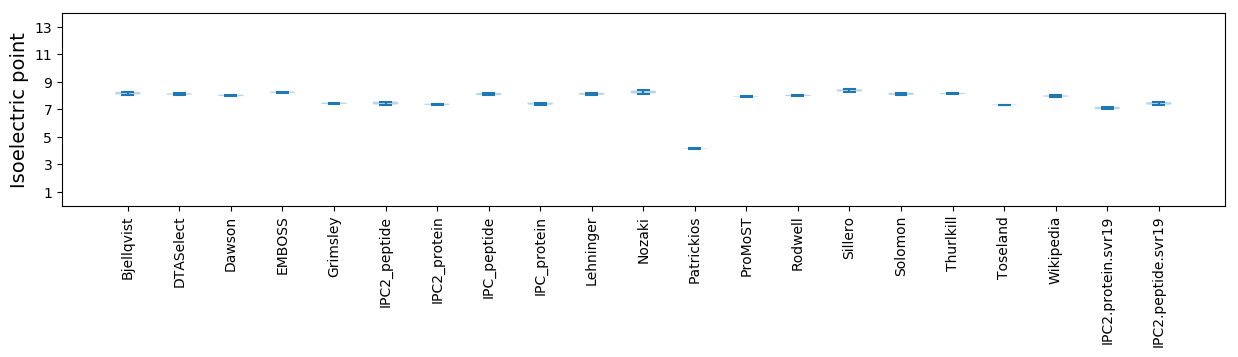
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A143BRM9|A0A143BRM9_9VIRU Coat protein OS=Rhizoctonia solani dsRNA virus 3 OX=1825688 PE=4 SV=1
MM1 pKa = 7.89SDD3 pKa = 3.57PAIKK7 pKa = 10.61SIDD10 pKa = 3.52DD11 pKa = 3.29VMSYY15 pKa = 8.75VRR17 pKa = 11.84KK18 pKa = 9.71QFEE21 pKa = 4.3EE22 pKa = 3.95MRR24 pKa = 11.84SFQQAATPPVPEE36 pKa = 4.39KK37 pKa = 11.01KK38 pKa = 9.58EE39 pKa = 3.81ARR41 pKa = 11.84DD42 pKa = 3.41ALGQRR47 pKa = 11.84DD48 pKa = 4.11VVAQAQKK55 pKa = 9.97ATTASASKK63 pKa = 10.54PPGPSSAPDD72 pKa = 3.26VPVPFVHH79 pKa = 7.07TSDD82 pKa = 3.68NVTNSLRR89 pKa = 11.84LALGLNFRR97 pKa = 11.84ATPVPQFGSNWYY109 pKa = 9.3LPSYY113 pKa = 9.95FLSHH117 pKa = 7.03QILDD121 pKa = 3.35ILNRR125 pKa = 11.84KK126 pKa = 7.98VFSVRR131 pKa = 11.84KK132 pKa = 9.01FYY134 pKa = 10.52EE135 pKa = 3.88ASEE138 pKa = 4.08FLDD141 pKa = 3.67PLLVHH146 pKa = 6.58VYY148 pKa = 10.01IQILEE153 pKa = 4.55IIHH156 pKa = 5.96TLRR159 pKa = 11.84AQQQGSTISTEE170 pKa = 4.04NEE172 pKa = 3.8LFLSWFDD179 pKa = 3.94NNFPSDD185 pKa = 3.87TLPVPGTHH193 pKa = 5.26VTLIANLAASSPAIGSYY210 pKa = 11.21SNIFPRR216 pKa = 11.84LPDD219 pKa = 3.9DD220 pKa = 4.75SRR222 pKa = 11.84ATTNTNGVPTTNAATGTMHH241 pKa = 6.92QLTGRR246 pKa = 11.84LVPIPGILDD255 pKa = 3.59VYY257 pKa = 9.52RR258 pKa = 11.84TRR260 pKa = 11.84LVLAINQINAQSPSTMTVGNFNLAHH285 pKa = 5.46RR286 pKa = 11.84TVLNLQCLTGAEE298 pKa = 4.09NSFTGPYY305 pKa = 8.51YY306 pKa = 9.62QAPVAAGLPATVAYY320 pKa = 9.59FSSPGVITAPWMSATAASSFMNNSLYY346 pKa = 10.1AQAMFGEE353 pKa = 4.98PSTANRR359 pKa = 11.84SEE361 pKa = 5.7DD362 pKa = 3.6FLSWQSFCGLDD373 pKa = 5.24GDD375 pKa = 4.63TQWFTEE381 pKa = 4.47TARR384 pKa = 11.84LMTVYY389 pKa = 10.51CKK391 pKa = 10.04FVKK394 pKa = 10.73GSAPLSAIPFSGHH407 pKa = 5.08MANHH411 pKa = 5.45VTWSPSSAPYY421 pKa = 10.5GPATHH426 pKa = 7.73RR427 pKa = 11.84YY428 pKa = 8.51RR429 pKa = 11.84EE430 pKa = 4.36RR431 pKa = 11.84ISMAGNGSVQPRR443 pKa = 11.84YY444 pKa = 9.3PRR446 pKa = 11.84SRR448 pKa = 11.84HH449 pKa = 4.66CC450 pKa = 4.62
MM1 pKa = 7.89SDD3 pKa = 3.57PAIKK7 pKa = 10.61SIDD10 pKa = 3.52DD11 pKa = 3.29VMSYY15 pKa = 8.75VRR17 pKa = 11.84KK18 pKa = 9.71QFEE21 pKa = 4.3EE22 pKa = 3.95MRR24 pKa = 11.84SFQQAATPPVPEE36 pKa = 4.39KK37 pKa = 11.01KK38 pKa = 9.58EE39 pKa = 3.81ARR41 pKa = 11.84DD42 pKa = 3.41ALGQRR47 pKa = 11.84DD48 pKa = 4.11VVAQAQKK55 pKa = 9.97ATTASASKK63 pKa = 10.54PPGPSSAPDD72 pKa = 3.26VPVPFVHH79 pKa = 7.07TSDD82 pKa = 3.68NVTNSLRR89 pKa = 11.84LALGLNFRR97 pKa = 11.84ATPVPQFGSNWYY109 pKa = 9.3LPSYY113 pKa = 9.95FLSHH117 pKa = 7.03QILDD121 pKa = 3.35ILNRR125 pKa = 11.84KK126 pKa = 7.98VFSVRR131 pKa = 11.84KK132 pKa = 9.01FYY134 pKa = 10.52EE135 pKa = 3.88ASEE138 pKa = 4.08FLDD141 pKa = 3.67PLLVHH146 pKa = 6.58VYY148 pKa = 10.01IQILEE153 pKa = 4.55IIHH156 pKa = 5.96TLRR159 pKa = 11.84AQQQGSTISTEE170 pKa = 4.04NEE172 pKa = 3.8LFLSWFDD179 pKa = 3.94NNFPSDD185 pKa = 3.87TLPVPGTHH193 pKa = 5.26VTLIANLAASSPAIGSYY210 pKa = 11.21SNIFPRR216 pKa = 11.84LPDD219 pKa = 3.9DD220 pKa = 4.75SRR222 pKa = 11.84ATTNTNGVPTTNAATGTMHH241 pKa = 6.92QLTGRR246 pKa = 11.84LVPIPGILDD255 pKa = 3.59VYY257 pKa = 9.52RR258 pKa = 11.84TRR260 pKa = 11.84LVLAINQINAQSPSTMTVGNFNLAHH285 pKa = 5.46RR286 pKa = 11.84TVLNLQCLTGAEE298 pKa = 4.09NSFTGPYY305 pKa = 8.51YY306 pKa = 9.62QAPVAAGLPATVAYY320 pKa = 9.59FSSPGVITAPWMSATAASSFMNNSLYY346 pKa = 10.1AQAMFGEE353 pKa = 4.98PSTANRR359 pKa = 11.84SEE361 pKa = 5.7DD362 pKa = 3.6FLSWQSFCGLDD373 pKa = 5.24GDD375 pKa = 4.63TQWFTEE381 pKa = 4.47TARR384 pKa = 11.84LMTVYY389 pKa = 10.51CKK391 pKa = 10.04FVKK394 pKa = 10.73GSAPLSAIPFSGHH407 pKa = 5.08MANHH411 pKa = 5.45VTWSPSSAPYY421 pKa = 10.5GPATHH426 pKa = 7.73RR427 pKa = 11.84YY428 pKa = 8.51RR429 pKa = 11.84EE430 pKa = 4.36RR431 pKa = 11.84ISMAGNGSVQPRR443 pKa = 11.84YY444 pKa = 9.3PRR446 pKa = 11.84SRR448 pKa = 11.84HH449 pKa = 4.66CC450 pKa = 4.62
Molecular weight: 49.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1032 |
450 |
582 |
516.0 |
58.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.752 ± 1.427 | 0.872 ± 0.011 |
5.62 ± 1.17 | 3.973 ± 0.547 |
6.202 ± 0.692 | 4.36 ± 0.618 |
3.295 ± 0.54 | 5.523 ± 0.826 |
3.779 ± 0.988 | 8.43 ± 0.414 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.907 ± 0.294 | 4.651 ± 0.433 |
6.783 ± 0.914 | 3.779 ± 0.563 |
6.202 ± 0.692 | 7.074 ± 1.999 |
5.911 ± 1.326 | 6.686 ± 0.153 |
1.647 ± 0.199 | 4.554 ± 0.775 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
