
Chicken stool associated circular virus 2
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.07
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A346BLF0|A0A346BLF0_9VIRU Capsid protein OS=Chicken stool associated circular virus 2 OX=2109374 PE=4 SV=1
MM1 pKa = 7.94RR2 pKa = 11.84SNFVISNSVANFRR15 pKa = 11.84ARR17 pKa = 11.84RR18 pKa = 11.84DD19 pKa = 3.05IEE21 pKa = 4.04TAAKK25 pKa = 9.75RR26 pKa = 11.84WCFTINNPTDD36 pKa = 3.57RR37 pKa = 11.84DD38 pKa = 4.12RR39 pKa = 11.84FWEE42 pKa = 4.38SPDD45 pKa = 3.59ALEE48 pKa = 3.8QLSYY52 pKa = 10.77IIVQEE57 pKa = 3.92EE58 pKa = 3.92RR59 pKa = 11.84GEE61 pKa = 4.23NGTVHH66 pKa = 5.51YY67 pKa = 10.46QGFLILKK74 pKa = 9.08KK75 pKa = 10.1RR76 pKa = 11.84NRR78 pKa = 11.84VTWLKK83 pKa = 11.11RR84 pKa = 11.84NMNEE88 pKa = 3.56RR89 pKa = 11.84AHH91 pKa = 5.82WEE93 pKa = 4.07KK94 pKa = 10.25TRR96 pKa = 11.84GTDD99 pKa = 3.39KK100 pKa = 10.91QAADD104 pKa = 3.9YY105 pKa = 10.55CRR107 pKa = 11.84KK108 pKa = 10.24DD109 pKa = 3.56DD110 pKa = 3.53THH112 pKa = 8.26VPGGLRR118 pKa = 11.84FEE120 pKa = 4.79HH121 pKa = 6.37GKK123 pKa = 9.44PPRR126 pKa = 11.84EE127 pKa = 4.14KK128 pKa = 10.17KK129 pKa = 10.55DD130 pKa = 3.35KK131 pKa = 9.81EE132 pKa = 4.04QEE134 pKa = 3.93ANEE137 pKa = 4.13ILDD140 pKa = 3.9SLRR143 pKa = 11.84TGYY146 pKa = 10.13KK147 pKa = 9.75RR148 pKa = 11.84PQEE151 pKa = 4.08IPAEE155 pKa = 4.11LLRR158 pKa = 11.84SRR160 pKa = 11.84NFISAYY166 pKa = 9.9KK167 pKa = 10.67LITTDD172 pKa = 2.79VLGPFRR178 pKa = 11.84PNLKK182 pKa = 9.92IITLVGPPGCGKK194 pKa = 10.54SYY196 pKa = 10.46LINYY200 pKa = 7.58LFPDD204 pKa = 4.32CARR207 pKa = 11.84LIMGNAGFWWQNPCSKK223 pKa = 10.09VACIEE228 pKa = 4.04EE229 pKa = 4.22FAGQIPLQKK238 pKa = 9.64MLNLLDD244 pKa = 5.27PYY246 pKa = 10.09PQSLEE251 pKa = 4.25VKK253 pKa = 10.22GGTRR257 pKa = 11.84PCLFEE262 pKa = 5.05VIFITSNSCPEE273 pKa = 3.2DD274 pKa = 3.02WYY276 pKa = 10.94RR277 pKa = 11.84NKK279 pKa = 10.07VPGIPEE285 pKa = 3.48EE286 pKa = 4.26AKK288 pKa = 10.72RR289 pKa = 11.84HH290 pKa = 5.5DD291 pKa = 4.61ALLALYY297 pKa = 10.01DD298 pKa = 3.62RR299 pKa = 11.84LGYY302 pKa = 10.07HH303 pKa = 6.7PAWHH307 pKa = 6.72HH308 pKa = 5.76GALEE312 pKa = 4.32RR313 pKa = 11.84TCGHH317 pKa = 5.91YY318 pKa = 10.71LEE320 pKa = 5.5PPQLGAITPEE330 pKa = 4.59WIQEE334 pKa = 3.92TRR336 pKa = 11.84KK337 pKa = 8.9WLLRR341 pKa = 11.84EE342 pKa = 3.71VRR344 pKa = 11.84RR345 pKa = 11.84ILEE348 pKa = 4.11MPEE351 pKa = 3.88EE352 pKa = 4.21QPAAAAAAAQPEE364 pKa = 4.3EE365 pKa = 4.54QPTQRR370 pKa = 11.84LEE372 pKa = 3.96EE373 pKa = 4.42AEE375 pKa = 4.64EE376 pKa = 4.36FPEE379 pKa = 4.17HH380 pKa = 6.37QPEE383 pKa = 4.3LTLLEE388 pKa = 4.56TTTSSQTSITLKK400 pKa = 10.59LSADD404 pKa = 3.43KK405 pKa = 10.06THH407 pKa = 6.97LGKK410 pKa = 9.23TSTSSKK416 pKa = 10.28HH417 pKa = 5.72KK418 pKa = 10.6AHH420 pKa = 6.27NTQDD424 pKa = 3.01THH426 pKa = 4.28STKK429 pKa = 10.61RR430 pKa = 11.84KK431 pKa = 8.89FVSQLRR437 pKa = 11.84QLKK440 pKa = 9.58LWAVV444 pKa = 3.6
MM1 pKa = 7.94RR2 pKa = 11.84SNFVISNSVANFRR15 pKa = 11.84ARR17 pKa = 11.84RR18 pKa = 11.84DD19 pKa = 3.05IEE21 pKa = 4.04TAAKK25 pKa = 9.75RR26 pKa = 11.84WCFTINNPTDD36 pKa = 3.57RR37 pKa = 11.84DD38 pKa = 4.12RR39 pKa = 11.84FWEE42 pKa = 4.38SPDD45 pKa = 3.59ALEE48 pKa = 3.8QLSYY52 pKa = 10.77IIVQEE57 pKa = 3.92EE58 pKa = 3.92RR59 pKa = 11.84GEE61 pKa = 4.23NGTVHH66 pKa = 5.51YY67 pKa = 10.46QGFLILKK74 pKa = 9.08KK75 pKa = 10.1RR76 pKa = 11.84NRR78 pKa = 11.84VTWLKK83 pKa = 11.11RR84 pKa = 11.84NMNEE88 pKa = 3.56RR89 pKa = 11.84AHH91 pKa = 5.82WEE93 pKa = 4.07KK94 pKa = 10.25TRR96 pKa = 11.84GTDD99 pKa = 3.39KK100 pKa = 10.91QAADD104 pKa = 3.9YY105 pKa = 10.55CRR107 pKa = 11.84KK108 pKa = 10.24DD109 pKa = 3.56DD110 pKa = 3.53THH112 pKa = 8.26VPGGLRR118 pKa = 11.84FEE120 pKa = 4.79HH121 pKa = 6.37GKK123 pKa = 9.44PPRR126 pKa = 11.84EE127 pKa = 4.14KK128 pKa = 10.17KK129 pKa = 10.55DD130 pKa = 3.35KK131 pKa = 9.81EE132 pKa = 4.04QEE134 pKa = 3.93ANEE137 pKa = 4.13ILDD140 pKa = 3.9SLRR143 pKa = 11.84TGYY146 pKa = 10.13KK147 pKa = 9.75RR148 pKa = 11.84PQEE151 pKa = 4.08IPAEE155 pKa = 4.11LLRR158 pKa = 11.84SRR160 pKa = 11.84NFISAYY166 pKa = 9.9KK167 pKa = 10.67LITTDD172 pKa = 2.79VLGPFRR178 pKa = 11.84PNLKK182 pKa = 9.92IITLVGPPGCGKK194 pKa = 10.54SYY196 pKa = 10.46LINYY200 pKa = 7.58LFPDD204 pKa = 4.32CARR207 pKa = 11.84LIMGNAGFWWQNPCSKK223 pKa = 10.09VACIEE228 pKa = 4.04EE229 pKa = 4.22FAGQIPLQKK238 pKa = 9.64MLNLLDD244 pKa = 5.27PYY246 pKa = 10.09PQSLEE251 pKa = 4.25VKK253 pKa = 10.22GGTRR257 pKa = 11.84PCLFEE262 pKa = 5.05VIFITSNSCPEE273 pKa = 3.2DD274 pKa = 3.02WYY276 pKa = 10.94RR277 pKa = 11.84NKK279 pKa = 10.07VPGIPEE285 pKa = 3.48EE286 pKa = 4.26AKK288 pKa = 10.72RR289 pKa = 11.84HH290 pKa = 5.5DD291 pKa = 4.61ALLALYY297 pKa = 10.01DD298 pKa = 3.62RR299 pKa = 11.84LGYY302 pKa = 10.07HH303 pKa = 6.7PAWHH307 pKa = 6.72HH308 pKa = 5.76GALEE312 pKa = 4.32RR313 pKa = 11.84TCGHH317 pKa = 5.91YY318 pKa = 10.71LEE320 pKa = 5.5PPQLGAITPEE330 pKa = 4.59WIQEE334 pKa = 3.92TRR336 pKa = 11.84KK337 pKa = 8.9WLLRR341 pKa = 11.84EE342 pKa = 3.71VRR344 pKa = 11.84RR345 pKa = 11.84ILEE348 pKa = 4.11MPEE351 pKa = 3.88EE352 pKa = 4.21QPAAAAAAAQPEE364 pKa = 4.3EE365 pKa = 4.54QPTQRR370 pKa = 11.84LEE372 pKa = 3.96EE373 pKa = 4.42AEE375 pKa = 4.64EE376 pKa = 4.36FPEE379 pKa = 4.17HH380 pKa = 6.37QPEE383 pKa = 4.3LTLLEE388 pKa = 4.56TTTSSQTSITLKK400 pKa = 10.59LSADD404 pKa = 3.43KK405 pKa = 10.06THH407 pKa = 6.97LGKK410 pKa = 9.23TSTSSKK416 pKa = 10.28HH417 pKa = 5.72KK418 pKa = 10.6AHH420 pKa = 6.27NTQDD424 pKa = 3.01THH426 pKa = 4.28STKK429 pKa = 10.61RR430 pKa = 11.84KK431 pKa = 8.89FVSQLRR437 pKa = 11.84QLKK440 pKa = 9.58LWAVV444 pKa = 3.6
Molecular weight: 51.13 kDa
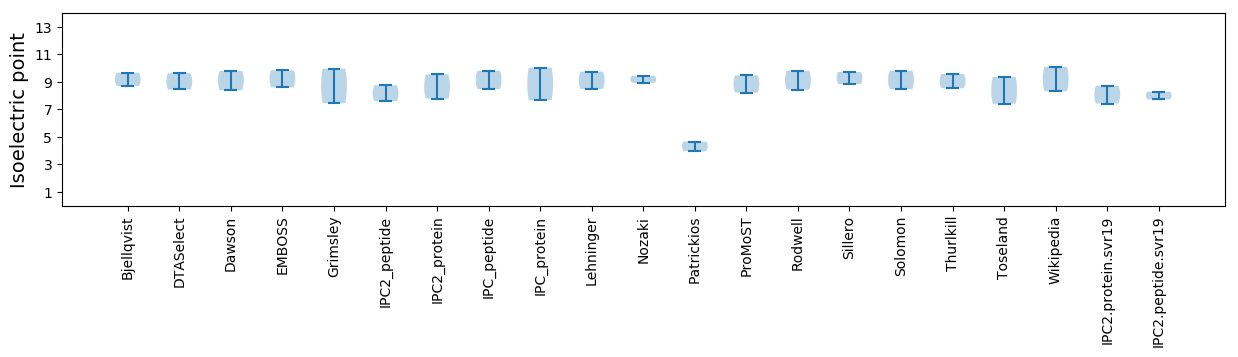
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BLF0|A0A346BLF0_9VIRU Capsid protein OS=Chicken stool associated circular virus 2 OX=2109374 PE=4 SV=1
MM1 pKa = 7.6PRR3 pKa = 11.84YY4 pKa = 9.49KK5 pKa = 10.32SRR7 pKa = 11.84RR8 pKa = 11.84TSSRR12 pKa = 11.84YY13 pKa = 6.91SRR15 pKa = 11.84RR16 pKa = 11.84YY17 pKa = 8.52RR18 pKa = 11.84RR19 pKa = 11.84YY20 pKa = 8.67RR21 pKa = 11.84RR22 pKa = 11.84YY23 pKa = 9.88RR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 9.85ARR28 pKa = 11.84IPRR31 pKa = 11.84RR32 pKa = 11.84VSYY35 pKa = 11.52ANGRR39 pKa = 11.84DD40 pKa = 3.48SSRR43 pKa = 11.84TNIKK47 pKa = 10.23VVTTTYY53 pKa = 10.57RR54 pKa = 11.84AGDD57 pKa = 3.46IAAGANDD64 pKa = 4.55SIVFTLPAFSRR75 pKa = 11.84TGGSGPSIVDD85 pKa = 3.77SICVLNSPLYY95 pKa = 9.59RR96 pKa = 11.84AYY98 pKa = 9.26TSLYY102 pKa = 10.63EE103 pKa = 4.07EE104 pKa = 4.17VLLRR108 pKa = 11.84GVSYY112 pKa = 10.55EE113 pKa = 3.75ITVNNPIGASATTLPPFMIFTSVDD137 pKa = 2.46RR138 pKa = 11.84RR139 pKa = 11.84YY140 pKa = 10.72AYY142 pKa = 10.61GEE144 pKa = 4.13VPPSQSQLTNYY155 pKa = 10.07SSSKK159 pKa = 8.56PISYY163 pKa = 8.03VTYY166 pKa = 10.41NVTKK170 pKa = 9.18FRR172 pKa = 11.84KK173 pKa = 9.16YY174 pKa = 9.91VGARR178 pKa = 11.84DD179 pKa = 3.82LVEE182 pKa = 3.63KK183 pKa = 8.91TQYY186 pKa = 10.64HH187 pKa = 7.25DD188 pKa = 3.83CTVSYY193 pKa = 10.56YY194 pKa = 11.1NSGGNQYY201 pKa = 10.16WYY203 pKa = 10.51DD204 pKa = 3.59PAVNSAGVNPNFFNPALFFFVRR226 pKa = 11.84FPFAPTSTMQLQLSIRR242 pKa = 11.84CTFYY246 pKa = 10.53LTFRR250 pKa = 11.84NPAPFDD256 pKa = 3.94SVPVPAAAAAAASGPFEE273 pKa = 4.28SFSTLSADD281 pKa = 3.9EE282 pKa = 4.12QLLDD286 pKa = 4.46IPDD289 pKa = 3.97PHH291 pKa = 7.0PLSGSTDD298 pKa = 3.42PPLIEE303 pKa = 4.13TT304 pKa = 4.52
MM1 pKa = 7.6PRR3 pKa = 11.84YY4 pKa = 9.49KK5 pKa = 10.32SRR7 pKa = 11.84RR8 pKa = 11.84TSSRR12 pKa = 11.84YY13 pKa = 6.91SRR15 pKa = 11.84RR16 pKa = 11.84YY17 pKa = 8.52RR18 pKa = 11.84RR19 pKa = 11.84YY20 pKa = 8.67RR21 pKa = 11.84RR22 pKa = 11.84YY23 pKa = 9.88RR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 9.85ARR28 pKa = 11.84IPRR31 pKa = 11.84RR32 pKa = 11.84VSYY35 pKa = 11.52ANGRR39 pKa = 11.84DD40 pKa = 3.48SSRR43 pKa = 11.84TNIKK47 pKa = 10.23VVTTTYY53 pKa = 10.57RR54 pKa = 11.84AGDD57 pKa = 3.46IAAGANDD64 pKa = 4.55SIVFTLPAFSRR75 pKa = 11.84TGGSGPSIVDD85 pKa = 3.77SICVLNSPLYY95 pKa = 9.59RR96 pKa = 11.84AYY98 pKa = 9.26TSLYY102 pKa = 10.63EE103 pKa = 4.07EE104 pKa = 4.17VLLRR108 pKa = 11.84GVSYY112 pKa = 10.55EE113 pKa = 3.75ITVNNPIGASATTLPPFMIFTSVDD137 pKa = 2.46RR138 pKa = 11.84RR139 pKa = 11.84YY140 pKa = 10.72AYY142 pKa = 10.61GEE144 pKa = 4.13VPPSQSQLTNYY155 pKa = 10.07SSSKK159 pKa = 8.56PISYY163 pKa = 8.03VTYY166 pKa = 10.41NVTKK170 pKa = 9.18FRR172 pKa = 11.84KK173 pKa = 9.16YY174 pKa = 9.91VGARR178 pKa = 11.84DD179 pKa = 3.82LVEE182 pKa = 3.63KK183 pKa = 8.91TQYY186 pKa = 10.64HH187 pKa = 7.25DD188 pKa = 3.83CTVSYY193 pKa = 10.56YY194 pKa = 11.1NSGGNQYY201 pKa = 10.16WYY203 pKa = 10.51DD204 pKa = 3.59PAVNSAGVNPNFFNPALFFFVRR226 pKa = 11.84FPFAPTSTMQLQLSIRR242 pKa = 11.84CTFYY246 pKa = 10.53LTFRR250 pKa = 11.84NPAPFDD256 pKa = 3.94SVPVPAAAAAAASGPFEE273 pKa = 4.28SFSTLSADD281 pKa = 3.9EE282 pKa = 4.12QLLDD286 pKa = 4.46IPDD289 pKa = 3.97PHH291 pKa = 7.0PLSGSTDD298 pKa = 3.42PPLIEE303 pKa = 4.13TT304 pKa = 4.52
Molecular weight: 34.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
748 |
304 |
444 |
374.0 |
42.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.62 ± 0.36 | 1.604 ± 0.368 |
4.144 ± 0.079 | 6.283 ± 2.177 |
4.278 ± 0.783 | 5.08 ± 0.087 |
2.139 ± 0.883 | 4.813 ± 0.32 |
4.679 ± 1.613 | 8.155 ± 1.332 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.07 ± 0.049 | 4.545 ± 0.232 |
7.487 ± 0.635 | 3.743 ± 0.859 |
8.289 ± 0.549 | 7.62 ± 2.32 |
7.219 ± 0.403 | 4.813 ± 1.249 |
1.604 ± 0.76 | 4.813 ± 1.837 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
