
Candidatus Marinamargulisbacteria bacterium SCGC AG-439-L15
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Candidatus Margulisbacteria; Candidatus Marinamargulisbacteria
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
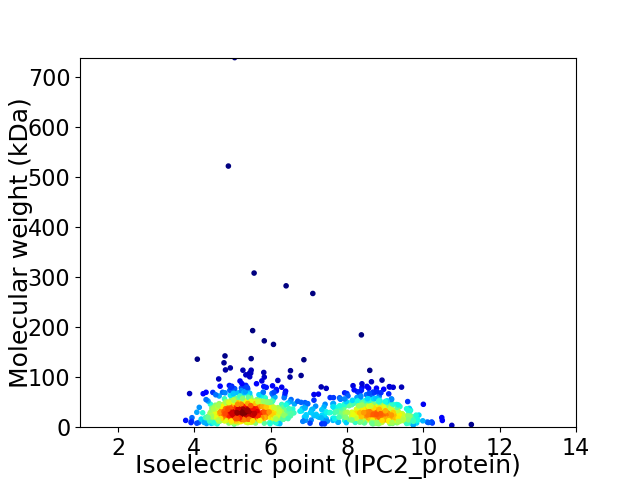
Virtual 2D-PAGE plot for 955 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A328RF04|A0A328RF04_9BACT Exodeoxyribonuclease III OS=Candidatus Marinamargulisbacteria bacterium SCGC AG-439-L15 OX=2184346 GN=xth PE=3 SV=1
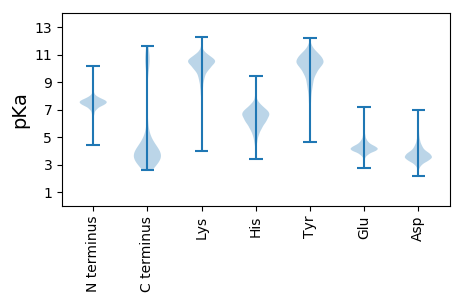
MM1 pKa = 7.51YY2 pKa = 10.44SGNCKK7 pKa = 9.68RR8 pKa = 11.84GYY10 pKa = 10.29IIGLMIAVVMVLSPLFSYY28 pKa = 10.68GALTPGVEE36 pKa = 3.99YY37 pKa = 10.32RR38 pKa = 11.84VVLEE42 pKa = 4.4RR43 pKa = 11.84LTSDD47 pKa = 3.42GEE49 pKa = 4.42GAIFTHH55 pKa = 5.44TTVTADD61 pKa = 3.04ANGIIEE67 pKa = 4.29YY68 pKa = 10.85AFDD71 pKa = 4.12NVPNCDD77 pKa = 3.58GADD80 pKa = 3.36SANFLIVSVIKK91 pKa = 10.55EE92 pKa = 4.4SDD94 pKa = 2.97WLGYY98 pKa = 9.68VPNSIVTSTVTFNVTEE114 pKa = 4.19EE115 pKa = 4.29SMSVVPAPPEE125 pKa = 3.91NTDD128 pKa = 3.06VFLGLNALSKK138 pKa = 10.65VQTKK142 pKa = 10.45GIIRR146 pKa = 11.84SFKK149 pKa = 9.28VTGKK153 pKa = 10.47ADD155 pKa = 3.73PLIALIGMVVFRR167 pKa = 11.84DD168 pKa = 3.73PDD170 pKa = 3.55LSDD173 pKa = 4.65SDD175 pKa = 3.64IQKK178 pKa = 10.09ISLGIADD185 pKa = 3.95QIFYY189 pKa = 10.15PGEE192 pKa = 3.74GFYY195 pKa = 11.1AQLSAVGVSDD205 pKa = 3.83EE206 pKa = 5.46KK207 pKa = 10.93IATFQKK213 pKa = 11.2ALVCEE218 pKa = 4.29SFGAGEE224 pKa = 4.1EE225 pKa = 4.39SFVGYY230 pKa = 8.9MRR232 pKa = 11.84EE233 pKa = 4.32TYY235 pKa = 10.49DD236 pKa = 3.57SNSVIMAGGDD246 pKa = 3.0EE247 pKa = 4.39GAARR251 pKa = 11.84EE252 pKa = 4.11AHH254 pKa = 5.91IKK256 pKa = 10.3AGEE259 pKa = 3.9RR260 pKa = 11.84LGAIVINAAEE270 pKa = 4.09YY271 pKa = 11.3AEE273 pKa = 4.81IDD275 pKa = 4.05MEE277 pKa = 4.67DD278 pKa = 3.75LLHH281 pKa = 6.89AVEE284 pKa = 5.3SLDD287 pKa = 5.42FEE289 pKa = 4.71DD290 pKa = 3.52TDD292 pKa = 4.65EE293 pKa = 4.2YY294 pKa = 11.65AFDD297 pKa = 3.74NRR299 pKa = 11.84IEE301 pKa = 4.0QLLSNSLSTFFQRR314 pKa = 11.84IYY316 pKa = 11.33VEE318 pKa = 4.59GTGSRR323 pKa = 11.84YY324 pKa = 8.28RR325 pKa = 11.84TALAGINASDD335 pKa = 3.65SQASRR340 pKa = 11.84FEE342 pKa = 4.68SAFSTLISSVEE353 pKa = 4.07EE354 pKa = 3.9LTDD357 pKa = 3.71PDD359 pKa = 3.51IYY361 pKa = 11.11EE362 pKa = 4.46FSDD365 pKa = 3.52SVAAFDD371 pKa = 5.54DD372 pKa = 4.43SRR374 pKa = 11.84DD375 pKa = 3.59SQYY378 pKa = 11.95DD379 pKa = 3.33SAQYY383 pKa = 10.42QFAVAIQSTEE393 pKa = 3.74AEE395 pKa = 4.19INALIDD401 pKa = 3.49TVADD405 pKa = 3.88IIGFDD410 pKa = 4.45DD411 pKa = 3.52PTSYY415 pKa = 11.0FNQNYY420 pKa = 9.98GSDD423 pKa = 3.49DD424 pKa = 3.42WGDD427 pKa = 3.28GRR429 pKa = 11.84TVSDD433 pKa = 3.83NANSHH438 pKa = 6.74WIFDD442 pKa = 4.25SLRR445 pKa = 11.84HH446 pKa = 6.76DD447 pKa = 3.7YY448 pKa = 9.64GTFFAEE454 pKa = 4.94IYY456 pKa = 10.49DD457 pKa = 4.2SGSGQYY463 pKa = 10.63NYY465 pKa = 10.41LRR467 pKa = 11.84ASIPQTVAFQYY478 pKa = 10.85VADD481 pKa = 4.32HH482 pKa = 6.19LQSGGTFSYY491 pKa = 9.52TWVDD495 pKa = 2.83IYY497 pKa = 11.28GGCDD501 pKa = 2.8TDD503 pKa = 4.37EE504 pKa = 4.62NVSSSDD510 pKa = 2.94ASMGMDD516 pKa = 4.51DD517 pKa = 3.27WWEE520 pKa = 4.47SNDD523 pKa = 3.32PTVWLWQLMDD533 pKa = 4.24LQWNVIDD540 pKa = 4.07LQRR543 pKa = 11.84WRR545 pKa = 11.84SDD547 pKa = 3.51YY548 pKa = 11.71YY549 pKa = 10.21NVQYY553 pKa = 11.08YY554 pKa = 10.35GADD557 pKa = 3.65DD558 pKa = 5.25FEE560 pKa = 4.58MTDD563 pKa = 2.83ITKK566 pKa = 10.42CYY568 pKa = 10.02STFLDD573 pKa = 3.69TMVTNINSDD582 pKa = 3.23MPAASKK588 pKa = 10.65KK589 pKa = 10.6YY590 pKa = 9.95ILNLFLRR597 pKa = 11.84PDD599 pKa = 3.41NNEE602 pKa = 4.13DD603 pKa = 3.62YY604 pKa = 11.38NPP606 pKa = 3.59
MM1 pKa = 7.51YY2 pKa = 10.44SGNCKK7 pKa = 9.68RR8 pKa = 11.84GYY10 pKa = 10.29IIGLMIAVVMVLSPLFSYY28 pKa = 10.68GALTPGVEE36 pKa = 3.99YY37 pKa = 10.32RR38 pKa = 11.84VVLEE42 pKa = 4.4RR43 pKa = 11.84LTSDD47 pKa = 3.42GEE49 pKa = 4.42GAIFTHH55 pKa = 5.44TTVTADD61 pKa = 3.04ANGIIEE67 pKa = 4.29YY68 pKa = 10.85AFDD71 pKa = 4.12NVPNCDD77 pKa = 3.58GADD80 pKa = 3.36SANFLIVSVIKK91 pKa = 10.55EE92 pKa = 4.4SDD94 pKa = 2.97WLGYY98 pKa = 9.68VPNSIVTSTVTFNVTEE114 pKa = 4.19EE115 pKa = 4.29SMSVVPAPPEE125 pKa = 3.91NTDD128 pKa = 3.06VFLGLNALSKK138 pKa = 10.65VQTKK142 pKa = 10.45GIIRR146 pKa = 11.84SFKK149 pKa = 9.28VTGKK153 pKa = 10.47ADD155 pKa = 3.73PLIALIGMVVFRR167 pKa = 11.84DD168 pKa = 3.73PDD170 pKa = 3.55LSDD173 pKa = 4.65SDD175 pKa = 3.64IQKK178 pKa = 10.09ISLGIADD185 pKa = 3.95QIFYY189 pKa = 10.15PGEE192 pKa = 3.74GFYY195 pKa = 11.1AQLSAVGVSDD205 pKa = 3.83EE206 pKa = 5.46KK207 pKa = 10.93IATFQKK213 pKa = 11.2ALVCEE218 pKa = 4.29SFGAGEE224 pKa = 4.1EE225 pKa = 4.39SFVGYY230 pKa = 8.9MRR232 pKa = 11.84EE233 pKa = 4.32TYY235 pKa = 10.49DD236 pKa = 3.57SNSVIMAGGDD246 pKa = 3.0EE247 pKa = 4.39GAARR251 pKa = 11.84EE252 pKa = 4.11AHH254 pKa = 5.91IKK256 pKa = 10.3AGEE259 pKa = 3.9RR260 pKa = 11.84LGAIVINAAEE270 pKa = 4.09YY271 pKa = 11.3AEE273 pKa = 4.81IDD275 pKa = 4.05MEE277 pKa = 4.67DD278 pKa = 3.75LLHH281 pKa = 6.89AVEE284 pKa = 5.3SLDD287 pKa = 5.42FEE289 pKa = 4.71DD290 pKa = 3.52TDD292 pKa = 4.65EE293 pKa = 4.2YY294 pKa = 11.65AFDD297 pKa = 3.74NRR299 pKa = 11.84IEE301 pKa = 4.0QLLSNSLSTFFQRR314 pKa = 11.84IYY316 pKa = 11.33VEE318 pKa = 4.59GTGSRR323 pKa = 11.84YY324 pKa = 8.28RR325 pKa = 11.84TALAGINASDD335 pKa = 3.65SQASRR340 pKa = 11.84FEE342 pKa = 4.68SAFSTLISSVEE353 pKa = 4.07EE354 pKa = 3.9LTDD357 pKa = 3.71PDD359 pKa = 3.51IYY361 pKa = 11.11EE362 pKa = 4.46FSDD365 pKa = 3.52SVAAFDD371 pKa = 5.54DD372 pKa = 4.43SRR374 pKa = 11.84DD375 pKa = 3.59SQYY378 pKa = 11.95DD379 pKa = 3.33SAQYY383 pKa = 10.42QFAVAIQSTEE393 pKa = 3.74AEE395 pKa = 4.19INALIDD401 pKa = 3.49TVADD405 pKa = 3.88IIGFDD410 pKa = 4.45DD411 pKa = 3.52PTSYY415 pKa = 11.0FNQNYY420 pKa = 9.98GSDD423 pKa = 3.49DD424 pKa = 3.42WGDD427 pKa = 3.28GRR429 pKa = 11.84TVSDD433 pKa = 3.83NANSHH438 pKa = 6.74WIFDD442 pKa = 4.25SLRR445 pKa = 11.84HH446 pKa = 6.76DD447 pKa = 3.7YY448 pKa = 9.64GTFFAEE454 pKa = 4.94IYY456 pKa = 10.49DD457 pKa = 4.2SGSGQYY463 pKa = 10.63NYY465 pKa = 10.41LRR467 pKa = 11.84ASIPQTVAFQYY478 pKa = 10.85VADD481 pKa = 4.32HH482 pKa = 6.19LQSGGTFSYY491 pKa = 9.52TWVDD495 pKa = 2.83IYY497 pKa = 11.28GGCDD501 pKa = 2.8TDD503 pKa = 4.37EE504 pKa = 4.62NVSSSDD510 pKa = 2.94ASMGMDD516 pKa = 4.51DD517 pKa = 3.27WWEE520 pKa = 4.47SNDD523 pKa = 3.32PTVWLWQLMDD533 pKa = 4.24LQWNVIDD540 pKa = 4.07LQRR543 pKa = 11.84WRR545 pKa = 11.84SDD547 pKa = 3.51YY548 pKa = 11.71YY549 pKa = 10.21NVQYY553 pKa = 11.08YY554 pKa = 10.35GADD557 pKa = 3.65DD558 pKa = 5.25FEE560 pKa = 4.58MTDD563 pKa = 2.83ITKK566 pKa = 10.42CYY568 pKa = 10.02STFLDD573 pKa = 3.69TMVTNINSDD582 pKa = 3.23MPAASKK588 pKa = 10.65KK589 pKa = 10.6YY590 pKa = 9.95ILNLFLRR597 pKa = 11.84PDD599 pKa = 3.41NNEE602 pKa = 4.13DD603 pKa = 3.62YY604 pKa = 11.38NPP606 pKa = 3.59
Molecular weight: 67.16 kDa
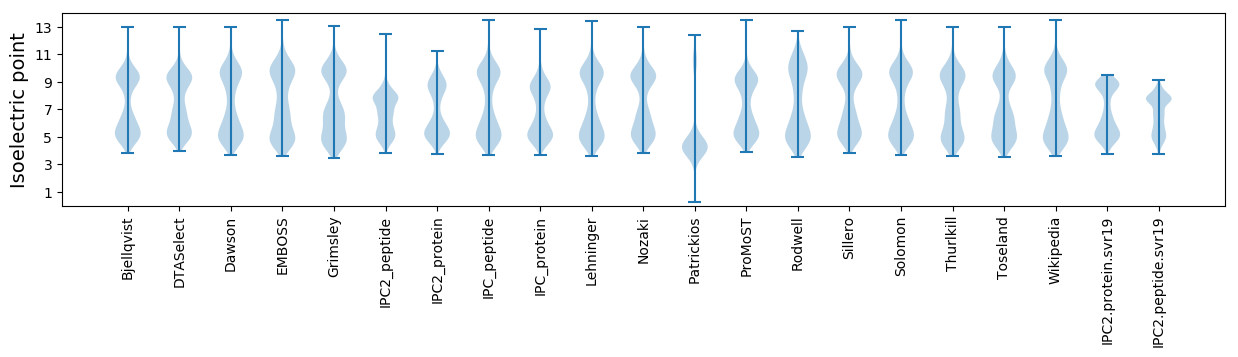
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A328RNM3|A0A328RNM3_9BACT Uncharacterized protein OS=Candidatus Marinamargulisbacteria bacterium SCGC AG-439-L15 OX=2184346 GN=DID77_03520 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.56RR3 pKa = 11.84TFQPNQRR10 pKa = 11.84KK11 pKa = 9.13RR12 pKa = 11.84KK13 pKa = 7.04KK14 pKa = 7.44THH16 pKa = 5.41GFLVRR21 pKa = 11.84MRR23 pKa = 11.84SVGGRR28 pKa = 11.84RR29 pKa = 11.84VIKK32 pKa = 9.82NRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.57GRR39 pKa = 11.84HH40 pKa = 3.82QLAAA44 pKa = 4.03
MM1 pKa = 7.44KK2 pKa = 9.56RR3 pKa = 11.84TFQPNQRR10 pKa = 11.84KK11 pKa = 9.13RR12 pKa = 11.84KK13 pKa = 7.04KK14 pKa = 7.44THH16 pKa = 5.41GFLVRR21 pKa = 11.84MRR23 pKa = 11.84SVGGRR28 pKa = 11.84RR29 pKa = 11.84VIKK32 pKa = 9.82NRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.57GRR39 pKa = 11.84HH40 pKa = 3.82QLAAA44 pKa = 4.03
Molecular weight: 5.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
321027 |
29 |
6683 |
336.2 |
37.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.393 ± 0.093 | 1.135 ± 0.038 |
5.538 ± 0.069 | 6.271 ± 0.075 |
4.517 ± 0.061 | 6.335 ± 0.088 |
2.197 ± 0.035 | 7.041 ± 0.09 |
7.411 ± 0.09 | 10.598 ± 0.079 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.18 ± 0.034 | 4.164 ± 0.053 |
4.004 ± 0.047 | 4.116 ± 0.068 |
3.966 ± 0.049 | 7.687 ± 0.085 |
5.952 ± 0.075 | 6.233 ± 0.076 |
0.841 ± 0.024 | 3.42 ± 0.051 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
