
Kordia sp. SMS9
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Kordia; unclassified Kordia
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
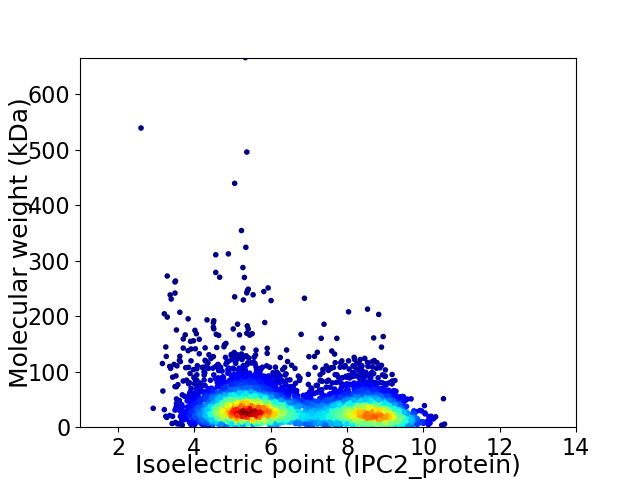
Virtual 2D-PAGE plot for 4631 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345H667|A0A345H667_9FLAO Uncharacterized protein OS=Kordia sp. SMS9 OX=2282170 GN=KORDIASMS9_04317 PE=4 SV=1
MM1 pKa = 7.28NKK3 pKa = 10.02KK4 pKa = 10.26LLFVLLFLTGVTTNFAQGIFDD25 pKa = 4.85DD26 pKa = 5.16SIHH29 pKa = 6.12TATTFGSVNSPGGEE43 pKa = 3.47AVQNVIDD50 pKa = 3.97QNSNTKK56 pKa = 10.2FLDD59 pKa = 3.72FNAFDD64 pKa = 5.04GIGFEE69 pKa = 4.12VDD71 pKa = 2.7ILGVSSTAVAMEE83 pKa = 4.22FVTANDD89 pKa = 3.45APEE92 pKa = 4.7RR93 pKa = 11.84DD94 pKa = 3.57PTNYY98 pKa = 9.97EE99 pKa = 3.47ISGSNDD105 pKa = 2.43GVNFTSIATGTIPCVSTRR123 pKa = 11.84FFSRR127 pKa = 11.84TFSFTNTTSYY137 pKa = 10.39TSYY140 pKa = 10.97RR141 pKa = 11.84LNFTGTCGASSINQIADD158 pKa = 3.31VQLFEE163 pKa = 5.6SIGDD167 pKa = 3.96SPVITCPSNITVNNTTGQCDD187 pKa = 3.8AIVNYY192 pKa = 9.15TVTANDD198 pKa = 4.09TEE200 pKa = 5.54DD201 pKa = 4.68GMLTPNVTSGLPSGASFPVGITEE224 pKa = 4.04VFMNVTDD231 pKa = 3.85TDD233 pKa = 4.29NNTSSCSFTVTVVDD247 pKa = 4.53NEE249 pKa = 4.57DD250 pKa = 3.67PVAGCPANIMQTAANFGDD268 pKa = 3.57TTAVVNYY275 pKa = 8.32TLSPTDD281 pKa = 3.24NCVLINPLTDD291 pKa = 4.09FTPLTNINGKK301 pKa = 10.0AYY303 pKa = 10.02YY304 pKa = 10.25LSNNVFTPANAFLDD318 pKa = 4.91AISQGGFVGTIRR330 pKa = 11.84NASDD334 pKa = 2.94NTAITEE340 pKa = 5.02AIATAGGAADD350 pKa = 3.74VLIGFSDD357 pKa = 3.72TDD359 pKa = 3.68VEE361 pKa = 5.24GTFVWQSGDD370 pKa = 3.54ASTFDD375 pKa = 3.04NWNPGEE381 pKa = 4.15PNNAGNEE388 pKa = 4.33DD389 pKa = 3.83YY390 pKa = 10.93TVIQSSGGWNDD401 pKa = 2.79ITNGSSYY408 pKa = 10.9RR409 pKa = 11.84YY410 pKa = 9.74LLEE413 pKa = 3.74VDD415 pKa = 3.89YY416 pKa = 11.31APTQTAGLPSGADD429 pKa = 3.54FPIGTTTNTFTITDD443 pKa = 4.09LAGNSVMCSFDD454 pKa = 3.27VTVDD458 pKa = 3.41ANLSVAEE465 pKa = 4.1NSFSNRR471 pKa = 11.84IKK473 pKa = 10.2LTPNPTKK480 pKa = 10.91GLISFSNTANISLEE494 pKa = 4.32SIVIMDD500 pKa = 4.25LNGRR504 pKa = 11.84ILQTVMNIGQLSDD517 pKa = 3.94GEE519 pKa = 4.7TIDD522 pKa = 3.86ISNFAAGMYY531 pKa = 8.94FVRR534 pKa = 11.84LQDD537 pKa = 3.29IEE539 pKa = 4.78NNIAVKK545 pKa = 10.45RR546 pKa = 11.84IIKK549 pKa = 9.37QQ550 pKa = 3.13
MM1 pKa = 7.28NKK3 pKa = 10.02KK4 pKa = 10.26LLFVLLFLTGVTTNFAQGIFDD25 pKa = 4.85DD26 pKa = 5.16SIHH29 pKa = 6.12TATTFGSVNSPGGEE43 pKa = 3.47AVQNVIDD50 pKa = 3.97QNSNTKK56 pKa = 10.2FLDD59 pKa = 3.72FNAFDD64 pKa = 5.04GIGFEE69 pKa = 4.12VDD71 pKa = 2.7ILGVSSTAVAMEE83 pKa = 4.22FVTANDD89 pKa = 3.45APEE92 pKa = 4.7RR93 pKa = 11.84DD94 pKa = 3.57PTNYY98 pKa = 9.97EE99 pKa = 3.47ISGSNDD105 pKa = 2.43GVNFTSIATGTIPCVSTRR123 pKa = 11.84FFSRR127 pKa = 11.84TFSFTNTTSYY137 pKa = 10.39TSYY140 pKa = 10.97RR141 pKa = 11.84LNFTGTCGASSINQIADD158 pKa = 3.31VQLFEE163 pKa = 5.6SIGDD167 pKa = 3.96SPVITCPSNITVNNTTGQCDD187 pKa = 3.8AIVNYY192 pKa = 9.15TVTANDD198 pKa = 4.09TEE200 pKa = 5.54DD201 pKa = 4.68GMLTPNVTSGLPSGASFPVGITEE224 pKa = 4.04VFMNVTDD231 pKa = 3.85TDD233 pKa = 4.29NNTSSCSFTVTVVDD247 pKa = 4.53NEE249 pKa = 4.57DD250 pKa = 3.67PVAGCPANIMQTAANFGDD268 pKa = 3.57TTAVVNYY275 pKa = 8.32TLSPTDD281 pKa = 3.24NCVLINPLTDD291 pKa = 4.09FTPLTNINGKK301 pKa = 10.0AYY303 pKa = 10.02YY304 pKa = 10.25LSNNVFTPANAFLDD318 pKa = 4.91AISQGGFVGTIRR330 pKa = 11.84NASDD334 pKa = 2.94NTAITEE340 pKa = 5.02AIATAGGAADD350 pKa = 3.74VLIGFSDD357 pKa = 3.72TDD359 pKa = 3.68VEE361 pKa = 5.24GTFVWQSGDD370 pKa = 3.54ASTFDD375 pKa = 3.04NWNPGEE381 pKa = 4.15PNNAGNEE388 pKa = 4.33DD389 pKa = 3.83YY390 pKa = 10.93TVIQSSGGWNDD401 pKa = 2.79ITNGSSYY408 pKa = 10.9RR409 pKa = 11.84YY410 pKa = 9.74LLEE413 pKa = 3.74VDD415 pKa = 3.89YY416 pKa = 11.31APTQTAGLPSGADD429 pKa = 3.54FPIGTTTNTFTITDD443 pKa = 4.09LAGNSVMCSFDD454 pKa = 3.27VTVDD458 pKa = 3.41ANLSVAEE465 pKa = 4.1NSFSNRR471 pKa = 11.84IKK473 pKa = 10.2LTPNPTKK480 pKa = 10.91GLISFSNTANISLEE494 pKa = 4.32SIVIMDD500 pKa = 4.25LNGRR504 pKa = 11.84ILQTVMNIGQLSDD517 pKa = 3.94GEE519 pKa = 4.7TIDD522 pKa = 3.86ISNFAAGMYY531 pKa = 8.94FVRR534 pKa = 11.84LQDD537 pKa = 3.29IEE539 pKa = 4.78NNIAVKK545 pKa = 10.45RR546 pKa = 11.84IIKK549 pKa = 9.37QQ550 pKa = 3.13
Molecular weight: 58.31 kDa
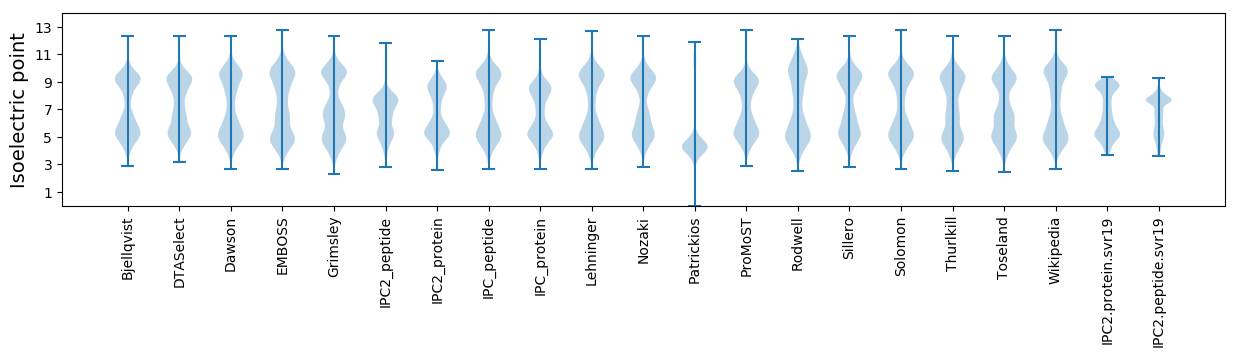
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345H787|A0A345H787_9FLAO Transcriptional activator NphR OS=Kordia sp. SMS9 OX=2282170 GN=nphR PE=4 SV=1
MM1 pKa = 7.32FPLVSAILASLFRR14 pKa = 11.84IIFHH18 pKa = 6.58NSITTIFKK26 pKa = 7.68THH28 pKa = 6.9HH29 pKa = 5.95FRR31 pKa = 11.84MHH33 pKa = 6.39LNISKK38 pKa = 7.08THH40 pKa = 6.35RR41 pKa = 11.84FLAIFSTFCIALLLARR57 pKa = 11.84ILIKK61 pKa = 10.86GSMFYY66 pKa = 11.17VFLVWNLFLAIIPYY80 pKa = 10.15AISSWLLHH88 pKa = 5.27TRR90 pKa = 11.84WIRR93 pKa = 11.84KK94 pKa = 7.33HH95 pKa = 5.62TFPLCIFLGIWLLFLPNAPYY115 pKa = 10.85LITDD119 pKa = 4.39LMHH122 pKa = 6.81LRR124 pKa = 11.84YY125 pKa = 9.97AKK127 pKa = 10.58SSVIWFDD134 pKa = 3.3ALLLFAFALNGLLLGIFSLQHH155 pKa = 4.73VFQVLMEE162 pKa = 4.47RR163 pKa = 11.84WNVKK167 pKa = 4.23TAKK170 pKa = 10.19RR171 pKa = 11.84ILFCVVFLCGFGMYY185 pKa = 10.56LGRR188 pKa = 11.84FLRR191 pKa = 11.84WNSWEE196 pKa = 4.23LFSDD200 pKa = 4.1PMLLAQDD207 pKa = 4.45CFRR210 pKa = 11.84SILYY214 pKa = 8.6PAYY217 pKa = 10.49RR218 pKa = 11.84MKK220 pKa = 10.18TWGITLGFGSFLWVLLLGVQTFIPIKK246 pKa = 10.22KK247 pKa = 8.56ATTII251 pKa = 3.6
MM1 pKa = 7.32FPLVSAILASLFRR14 pKa = 11.84IIFHH18 pKa = 6.58NSITTIFKK26 pKa = 7.68THH28 pKa = 6.9HH29 pKa = 5.95FRR31 pKa = 11.84MHH33 pKa = 6.39LNISKK38 pKa = 7.08THH40 pKa = 6.35RR41 pKa = 11.84FLAIFSTFCIALLLARR57 pKa = 11.84ILIKK61 pKa = 10.86GSMFYY66 pKa = 11.17VFLVWNLFLAIIPYY80 pKa = 10.15AISSWLLHH88 pKa = 5.27TRR90 pKa = 11.84WIRR93 pKa = 11.84KK94 pKa = 7.33HH95 pKa = 5.62TFPLCIFLGIWLLFLPNAPYY115 pKa = 10.85LITDD119 pKa = 4.39LMHH122 pKa = 6.81LRR124 pKa = 11.84YY125 pKa = 9.97AKK127 pKa = 10.58SSVIWFDD134 pKa = 3.3ALLLFAFALNGLLLGIFSLQHH155 pKa = 4.73VFQVLMEE162 pKa = 4.47RR163 pKa = 11.84WNVKK167 pKa = 4.23TAKK170 pKa = 10.19RR171 pKa = 11.84ILFCVVFLCGFGMYY185 pKa = 10.56LGRR188 pKa = 11.84FLRR191 pKa = 11.84WNSWEE196 pKa = 4.23LFSDD200 pKa = 4.1PMLLAQDD207 pKa = 4.45CFRR210 pKa = 11.84SILYY214 pKa = 8.6PAYY217 pKa = 10.49RR218 pKa = 11.84MKK220 pKa = 10.18TWGITLGFGSFLWVLLLGVQTFIPIKK246 pKa = 10.22KK247 pKa = 8.56ATTII251 pKa = 3.6
Molecular weight: 29.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1592471 |
29 |
5858 |
343.9 |
38.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.479 ± 0.035 | 0.848 ± 0.013 |
5.582 ± 0.049 | 6.568 ± 0.039 |
5.242 ± 0.032 | 5.961 ± 0.043 |
1.839 ± 0.017 | 7.938 ± 0.036 |
7.644 ± 0.059 | 8.991 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.091 ± 0.019 | 6.273 ± 0.046 |
3.268 ± 0.025 | 3.728 ± 0.026 |
3.385 ± 0.024 | 6.339 ± 0.029 |
6.545 ± 0.051 | 6.092 ± 0.029 |
1.002 ± 0.013 | 4.184 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
