
Thermoplasmatales archaeon SCGC AB-539-N05
Taxonomy: cellular organisms; Archaea; Candidatus Thermoplasmatota; Thermoplasmata; Thermoplasmatales; unclassified Thermoplasmatales
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
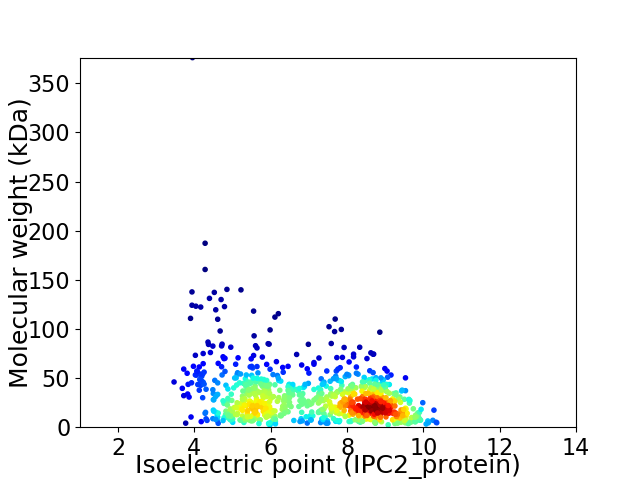
Virtual 2D-PAGE plot for 844 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M7TCL2|M7TCL2_9ARCH Uncharacterized protein OS=Thermoplasmatales archaeon SCGC AB-539-N05 OX=1198116 GN=MBGDN05_00153 PE=4 SV=1
DDD2 pKa = 4.32LSNEEE7 pKa = 3.94TKKK10 pKa = 9.43IIVSNVPPVANFTWTPTNPNQKKK33 pKa = 10.16DDD35 pKa = 3.48VSFNSTTSYYY45 pKa = 11.43DD46 pKa = 3.53DDD48 pKa = 3.69FIVNWSWTFGDDD60 pKa = 4.25NTSYYY65 pKa = 10.47EE66 pKa = 4.12NPSHHH71 pKa = 7.56YYY73 pKa = 10.31DD74 pKa = 3.71DD75 pKa = 4.11GNYYY79 pKa = 7.54ATLTIEEE86 pKa = 5.15DD87 pKa = 4.2DDD89 pKa = 4.23SHHH92 pKa = 5.56TINKKK97 pKa = 7.55IQVNNTLPTVDDD109 pKa = 6.87NFTWTATYYY118 pKa = 9.7FSHHH122 pKa = 6.97TDDD125 pKa = 3.25DDD127 pKa = 3.38VNFTDDD133 pKa = 3.61SSDDD137 pKa = 3.59DDD139 pKa = 3.83EEE141 pKa = 4.4IAWHHH146 pKa = 6.57EEE148 pKa = 3.76SDDD151 pKa = 3.43HHH153 pKa = 7.44SDDD156 pKa = 3.54EE157 pKa = 4.54NPLHHH162 pKa = 6.72FIDDD166 pKa = 3.64SVYYY170 pKa = 10.53VTLTVWDDD178 pKa = 4.8DD179 pKa = 3.72GASNSTSKKK188 pKa = 10.96VTVVNIPPTADDD200 pKa = 3.18TYYY203 pKa = 9.04PHHH206 pKa = 6.78FGIFEEE212 pKa = 4.9DD213 pKa = 3.7HHH215 pKa = 5.82TDDD218 pKa = 4.73STDDD222 pKa = 3.18DDD224 pKa = 4.04VIVNWTWNFGDDD236 pKa = 4.19NISYYY241 pKa = 8.45EE242 pKa = 3.97NPVYYY247 pKa = 10.61YYY249 pKa = 10.74KKK251 pKa = 10.81GNFTITLNVTDDD263 pKa = 5.63DD264 pKa = 4.77DD265 pKa = 4.82ATDDD269 pKa = 3.41ATTQNITVLNNPLVADDD286 pKa = 4.41TWNPTNPTDDD296 pKa = 4.31EEE298 pKa = 4.69VNFTDDD304 pKa = 3.7SKKK307 pKa = 11.38DD308 pKa = 3.09DDD310 pKa = 3.68TVVNWTWNFGDDD322 pKa = 4.38NHHH325 pKa = 6.84YYY327 pKa = 8.43QHHH330 pKa = 6.63THHH333 pKa = 6.55YYY335 pKa = 10.77DDD337 pKa = 3.25GTYYY341 pKa = 9.95VTLTVRR347 pKa = 11.84DDD349 pKa = 3.99NDDD352 pKa = 3.04EE353 pKa = 4.54NTTSKKK359 pKa = 11.07DD360 pKa = 3.07TVLNVPPVADDD371 pKa = 4.12TYYY374 pKa = 10.71PTTPTNTDDD383 pKa = 3.72IQFNDDD389 pKa = 3.51STDDD393 pKa = 3.1DDD395 pKa = 3.76TIVSWSWDDD404 pKa = 3.47GDDD407 pKa = 5.44NTSTNQNPSHHH418 pKa = 6.45YYY420 pKa = 10.2AIGNYYY426 pKa = 8.0VTLTVTDDD434 pKa = 3.66DDD436 pKa = 4.09ATDDD440 pKa = 4.06SQKKK444 pKa = 8.68VNVSASPVAKKK455 pKa = 10.3IYYY458 pKa = 10.41PIAYYY463 pKa = 9.48NVNEEE468 pKa = 4.21IKKK471 pKa = 10.32LDDD474 pKa = 3.36SYYY477 pKa = 11.32DD478 pKa = 3.33DDD480 pKa = 4.57YYY482 pKa = 10.69MNWSWNFGDDD492 pKa = 5.03NHHH495 pKa = 6.46YYY497 pKa = 11.0EE498 pKa = 4.45KK499 pKa = 10.79DD500 pKa = 3.61IHHH503 pKa = 7.43YYY505 pKa = 8.31AANTYYY511 pKa = 8.51VMLTVRR517 pKa = 11.84DDD519 pKa = 3.67SGATNSTSKKK529 pKa = 10.89DD530 pKa = 3.34IVTT
DDD2 pKa = 4.32LSNEEE7 pKa = 3.94TKKK10 pKa = 9.43IIVSNVPPVANFTWTPTNPNQKKK33 pKa = 10.16DDD35 pKa = 3.48VSFNSTTSYYY45 pKa = 11.43DD46 pKa = 3.53DDD48 pKa = 3.69FIVNWSWTFGDDD60 pKa = 4.25NTSYYY65 pKa = 10.47EE66 pKa = 4.12NPSHHH71 pKa = 7.56YYY73 pKa = 10.31DD74 pKa = 3.71DD75 pKa = 4.11GNYYY79 pKa = 7.54ATLTIEEE86 pKa = 5.15DD87 pKa = 4.2DDD89 pKa = 4.23SHHH92 pKa = 5.56TINKKK97 pKa = 7.55IQVNNTLPTVDDD109 pKa = 6.87NFTWTATYYY118 pKa = 9.7FSHHH122 pKa = 6.97TDDD125 pKa = 3.25DDD127 pKa = 3.38VNFTDDD133 pKa = 3.61SSDDD137 pKa = 3.59DDD139 pKa = 3.83EEE141 pKa = 4.4IAWHHH146 pKa = 6.57EEE148 pKa = 3.76SDDD151 pKa = 3.43HHH153 pKa = 7.44SDDD156 pKa = 3.54EE157 pKa = 4.54NPLHHH162 pKa = 6.72FIDDD166 pKa = 3.64SVYYY170 pKa = 10.53VTLTVWDDD178 pKa = 4.8DD179 pKa = 3.72GASNSTSKKK188 pKa = 10.96VTVVNIPPTADDD200 pKa = 3.18TYYY203 pKa = 9.04PHHH206 pKa = 6.78FGIFEEE212 pKa = 4.9DD213 pKa = 3.7HHH215 pKa = 5.82TDDD218 pKa = 4.73STDDD222 pKa = 3.18DDD224 pKa = 4.04VIVNWTWNFGDDD236 pKa = 4.19NISYYY241 pKa = 8.45EE242 pKa = 3.97NPVYYY247 pKa = 10.61YYY249 pKa = 10.74KKK251 pKa = 10.81GNFTITLNVTDDD263 pKa = 5.63DD264 pKa = 4.77DD265 pKa = 4.82ATDDD269 pKa = 3.41ATTQNITVLNNPLVADDD286 pKa = 4.41TWNPTNPTDDD296 pKa = 4.31EEE298 pKa = 4.69VNFTDDD304 pKa = 3.7SKKK307 pKa = 11.38DD308 pKa = 3.09DDD310 pKa = 3.68TVVNWTWNFGDDD322 pKa = 4.38NHHH325 pKa = 6.84YYY327 pKa = 8.43QHHH330 pKa = 6.63THHH333 pKa = 6.55YYY335 pKa = 10.77DDD337 pKa = 3.25GTYYY341 pKa = 9.95VTLTVRR347 pKa = 11.84DDD349 pKa = 3.99NDDD352 pKa = 3.04EE353 pKa = 4.54NTTSKKK359 pKa = 11.07DD360 pKa = 3.07TVLNVPPVADDD371 pKa = 4.12TYYY374 pKa = 10.71PTTPTNTDDD383 pKa = 3.72IQFNDDD389 pKa = 3.51STDDD393 pKa = 3.1DDD395 pKa = 3.76TIVSWSWDDD404 pKa = 3.47GDDD407 pKa = 5.44NTSTNQNPSHHH418 pKa = 6.45YYY420 pKa = 10.2AIGNYYY426 pKa = 8.0VTLTVTDDD434 pKa = 3.66DDD436 pKa = 4.09ATDDD440 pKa = 4.06SQKKK444 pKa = 8.68VNVSASPVAKKK455 pKa = 10.3IYYY458 pKa = 10.41PIAYYY463 pKa = 9.48NVNEEE468 pKa = 4.21IKKK471 pKa = 10.32LDDD474 pKa = 3.36SYYY477 pKa = 11.32DD478 pKa = 3.33DDD480 pKa = 4.57YYY482 pKa = 10.69MNWSWNFGDDD492 pKa = 5.03NHHH495 pKa = 6.46YYY497 pKa = 11.0EE498 pKa = 4.45KK499 pKa = 10.79DD500 pKa = 3.61IHHH503 pKa = 7.43YYY505 pKa = 8.31AANTYYY511 pKa = 8.51VMLTVRR517 pKa = 11.84DDD519 pKa = 3.67SGATNSTSKKK529 pKa = 10.89DD530 pKa = 3.34IVTT
Molecular weight: 59.15 kDa
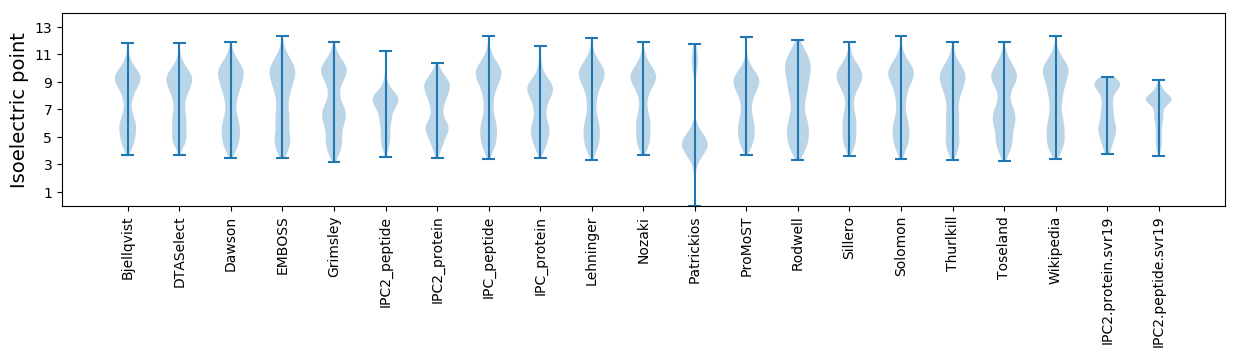
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M7U2Z1|M7U2Z1_9ARCH 30S ribosomal protein S2 OS=Thermoplasmatales archaeon SCGC AB-539-N05 OX=1198116 GN=rps2 PE=3 SV=1
QQ1 pKa = 6.66RR2 pKa = 11.84EE3 pKa = 3.63KK4 pKa = 10.7RR5 pKa = 11.84ARR7 pKa = 11.84TGAPMTYY14 pKa = 9.32TIHH17 pKa = 7.88DD18 pKa = 4.28KK19 pKa = 11.26GLSTTIGWQNRR30 pKa = 11.84DD31 pKa = 2.83AYY33 pKa = 10.37GRR35 pKa = 11.84SIPTRR40 pKa = 11.84NRR42 pKa = 11.84AQLYY46 pKa = 10.02RR47 pKa = 11.84LRR49 pKa = 11.84KK50 pKa = 6.68WQTRR54 pKa = 11.84TRR56 pKa = 11.84IGNATEE62 pKa = 4.03RR63 pKa = 11.84NLAIALSTIDD73 pKa = 3.91RR74 pKa = 11.84MSSGLSLPRR83 pKa = 11.84TVRR86 pKa = 11.84EE87 pKa = 4.05TAAMIYY93 pKa = 10.07RR94 pKa = 11.84KK95 pKa = 9.73AVLKK99 pKa = 10.63NLIRR103 pKa = 11.84GRR105 pKa = 11.84SVEE108 pKa = 4.15GVTAAALYY116 pKa = 8.71AACRR120 pKa = 11.84QCRR123 pKa = 11.84VPRR126 pKa = 11.84TLDD129 pKa = 3.74EE130 pKa = 4.4ISAIACMSRR139 pKa = 11.84KK140 pKa = 9.68EE141 pKa = 3.5IGRR144 pKa = 11.84NYY146 pKa = 10.15RR147 pKa = 11.84YY148 pKa = 9.68VARR151 pKa = 11.84EE152 pKa = 4.09LGLKK156 pKa = 10.12LMPTSPQDD164 pKa = 3.59YY165 pKa = 9.79ISRR168 pKa = 11.84FCSGLKK174 pKa = 10.52LSGDD178 pKa = 4.0VQAKK182 pKa = 8.31TLDD185 pKa = 3.63ILKK188 pKa = 9.99EE189 pKa = 3.85AADD192 pKa = 4.36RR193 pKa = 11.84EE194 pKa = 4.71LTSGRR199 pKa = 11.84GPTGIAAAALYY210 pKa = 9.97IATILCGEE218 pKa = 4.28RR219 pKa = 11.84RR220 pKa = 11.84TQRR223 pKa = 11.84EE224 pKa = 3.96VAEE227 pKa = 4.28VAGVTEE233 pKa = 3.89VTIRR237 pKa = 11.84NRR239 pKa = 11.84YY240 pKa = 9.01KK241 pKa = 10.51EE242 pKa = 4.18LSEE245 pKa = 4.18KK246 pKa = 10.87LDD248 pKa = 3.43INVILL253 pKa = 4.55
QQ1 pKa = 6.66RR2 pKa = 11.84EE3 pKa = 3.63KK4 pKa = 10.7RR5 pKa = 11.84ARR7 pKa = 11.84TGAPMTYY14 pKa = 9.32TIHH17 pKa = 7.88DD18 pKa = 4.28KK19 pKa = 11.26GLSTTIGWQNRR30 pKa = 11.84DD31 pKa = 2.83AYY33 pKa = 10.37GRR35 pKa = 11.84SIPTRR40 pKa = 11.84NRR42 pKa = 11.84AQLYY46 pKa = 10.02RR47 pKa = 11.84LRR49 pKa = 11.84KK50 pKa = 6.68WQTRR54 pKa = 11.84TRR56 pKa = 11.84IGNATEE62 pKa = 4.03RR63 pKa = 11.84NLAIALSTIDD73 pKa = 3.91RR74 pKa = 11.84MSSGLSLPRR83 pKa = 11.84TVRR86 pKa = 11.84EE87 pKa = 4.05TAAMIYY93 pKa = 10.07RR94 pKa = 11.84KK95 pKa = 9.73AVLKK99 pKa = 10.63NLIRR103 pKa = 11.84GRR105 pKa = 11.84SVEE108 pKa = 4.15GVTAAALYY116 pKa = 8.71AACRR120 pKa = 11.84QCRR123 pKa = 11.84VPRR126 pKa = 11.84TLDD129 pKa = 3.74EE130 pKa = 4.4ISAIACMSRR139 pKa = 11.84KK140 pKa = 9.68EE141 pKa = 3.5IGRR144 pKa = 11.84NYY146 pKa = 10.15RR147 pKa = 11.84YY148 pKa = 9.68VARR151 pKa = 11.84EE152 pKa = 4.09LGLKK156 pKa = 10.12LMPTSPQDD164 pKa = 3.59YY165 pKa = 9.79ISRR168 pKa = 11.84FCSGLKK174 pKa = 10.52LSGDD178 pKa = 4.0VQAKK182 pKa = 8.31TLDD185 pKa = 3.63ILKK188 pKa = 9.99EE189 pKa = 3.85AADD192 pKa = 4.36RR193 pKa = 11.84EE194 pKa = 4.71LTSGRR199 pKa = 11.84GPTGIAAAALYY210 pKa = 9.97IATILCGEE218 pKa = 4.28RR219 pKa = 11.84RR220 pKa = 11.84TQRR223 pKa = 11.84EE224 pKa = 3.96VAEE227 pKa = 4.28VAGVTEE233 pKa = 3.89VTIRR237 pKa = 11.84NRR239 pKa = 11.84YY240 pKa = 9.01KK241 pKa = 10.51EE242 pKa = 4.18LSEE245 pKa = 4.18KK246 pKa = 10.87LDD248 pKa = 3.43INVILL253 pKa = 4.55
Molecular weight: 28.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
238614 |
20 |
3387 |
282.7 |
31.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.05 ± 0.08 | 1.356 ± 0.035 |
5.926 ± 0.111 | 6.58 ± 0.089 |
4.237 ± 0.081 | 7.141 ± 0.083 |
1.854 ± 0.035 | 8.726 ± 0.107 |
8.273 ± 0.169 | 8.279 ± 0.145 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.452 ± 0.055 | 5.128 ± 0.122 |
3.863 ± 0.047 | 2.454 ± 0.044 |
3.855 ± 0.081 | 6.147 ± 0.127 |
5.78 ± 0.171 | 6.867 ± 0.084 |
1.251 ± 0.059 | 3.781 ± 0.084 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
