
Sanxia water strider virus 17
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
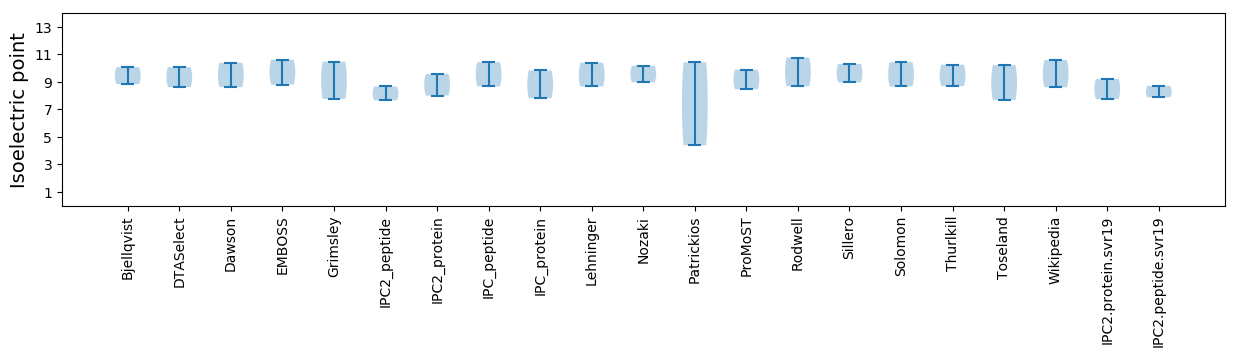
Average proteome isoelectric point is 8.48
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KG99|A0A1L3KG99_9VIRU Capsid protein OS=Sanxia water strider virus 17 OX=1923401 PE=3 SV=1
MM1 pKa = 8.03PINIRR6 pKa = 11.84TPVTASPEE14 pKa = 4.12QSSSHH19 pKa = 5.55TPFFCWKK26 pKa = 9.22VFNRR30 pKa = 11.84VQRR33 pKa = 11.84VTPKK37 pKa = 10.19PKK39 pKa = 9.9TSVATAEE46 pKa = 3.92YY47 pKa = 10.52RR48 pKa = 11.84PGKK51 pKa = 9.33VNLMQKK57 pKa = 8.45TRR59 pKa = 11.84DD60 pKa = 3.23RR61 pKa = 11.84FFKK64 pKa = 9.35MWNGNDD70 pKa = 3.11LPKK73 pKa = 10.14RR74 pKa = 11.84SKK76 pKa = 9.79YY77 pKa = 10.37ASNRR81 pKa = 11.84RR82 pKa = 11.84SLPPIDD88 pKa = 3.97FRR90 pKa = 11.84KK91 pKa = 9.3MLHH94 pKa = 6.13HH95 pKa = 6.68HH96 pKa = 6.92LKK98 pKa = 10.34SIPKK102 pKa = 9.37IPMASNHH109 pKa = 4.88THH111 pKa = 6.77PNAASLRR118 pKa = 11.84TSVAVEE124 pKa = 3.7LEE126 pKa = 4.21KK127 pKa = 10.69IVINAGFRR135 pKa = 11.84PYY137 pKa = 10.26SVSMSKK143 pKa = 10.01RR144 pKa = 11.84DD145 pKa = 3.42QYY147 pKa = 11.6DD148 pKa = 3.08GCRR151 pKa = 11.84YY152 pKa = 10.42YY153 pKa = 11.31FMDD156 pKa = 5.05KK157 pKa = 10.99DD158 pKa = 3.43LDD160 pKa = 3.57KK161 pKa = 11.35AFRR164 pKa = 11.84DD165 pKa = 3.6DD166 pKa = 4.31RR167 pKa = 11.84VTEE170 pKa = 3.97EE171 pKa = 4.66HH172 pKa = 6.53VLLMIDD178 pKa = 2.82VDD180 pKa = 4.4YY181 pKa = 11.62YY182 pKa = 11.7CDD184 pKa = 3.41INQYY188 pKa = 10.05LQLGNPIMIYY198 pKa = 9.57TFVPTEE204 pKa = 3.83AGGRR208 pKa = 11.84ALDD211 pKa = 3.93ASYY214 pKa = 10.07TINNNVVSYY223 pKa = 9.36CVKK226 pKa = 10.66GGATYY231 pKa = 10.1HH232 pKa = 6.41HH233 pKa = 6.84EE234 pKa = 3.88LWDD237 pKa = 3.81YY238 pKa = 11.37QGDD241 pKa = 3.96NVVVKK246 pKa = 10.3DD247 pKa = 3.39KK248 pKa = 11.23HH249 pKa = 6.68GNTLVYY255 pKa = 10.53VIEE258 pKa = 3.9QHH260 pKa = 7.18IIEE263 pKa = 4.84EE264 pKa = 4.4DD265 pKa = 3.28PNRR268 pKa = 11.84RR269 pKa = 11.84VIGFYY274 pKa = 10.49PIASYY279 pKa = 9.98PKK281 pKa = 8.19HH282 pKa = 5.41TCPFKK287 pKa = 10.42PEE289 pKa = 4.04SYY291 pKa = 10.76GFGRR295 pKa = 11.84LRR297 pKa = 11.84PSSMGVNCIRR307 pKa = 11.84NVTDD311 pKa = 5.07DD312 pKa = 4.23IISVAVQDD320 pKa = 3.69SANAVTIPQSIYY332 pKa = 10.02DD333 pKa = 3.74ALVVRR338 pKa = 11.84RR339 pKa = 11.84GEE341 pKa = 4.21SKK343 pKa = 10.83NPVIADD349 pKa = 3.17VEE351 pKa = 4.55RR352 pKa = 11.84ILNAEE357 pKa = 4.56HH358 pKa = 7.04IEE360 pKa = 4.04HH361 pKa = 6.47SAIKK365 pKa = 10.59APVLFKK371 pKa = 10.91LLQSSTTSCGVITSSATIGQAKK393 pKa = 9.74NFQTLFPLATEE404 pKa = 4.56DD405 pKa = 3.6GKK407 pKa = 10.26PVGRR411 pKa = 11.84AVAPSLVTDD420 pKa = 4.26PAFVPCKK427 pKa = 10.6SFNNDD432 pKa = 2.38NATIQGRR439 pKa = 11.84VNKK442 pKa = 9.42VRR444 pKa = 11.84NNNQSPPAWKK454 pKa = 8.95TYY456 pKa = 9.95DD457 pKa = 3.33AEE459 pKa = 4.27LVKK462 pKa = 10.6FIVPHH467 pKa = 6.03NKK469 pKa = 9.45NATGVPFDD477 pKa = 3.76YY478 pKa = 10.95EE479 pKa = 4.32RR480 pKa = 11.84VIEE483 pKa = 4.29LQNKK487 pKa = 6.24PAQRR491 pKa = 11.84GRR493 pKa = 11.84SEE495 pKa = 3.89QAKK498 pKa = 8.58ATVSNDD504 pKa = 3.03YY505 pKa = 10.54VNKK508 pKa = 9.83VKK510 pKa = 11.01AFIKK514 pKa = 10.36AEE516 pKa = 4.2AYY518 pKa = 10.44ASVTDD523 pKa = 3.92PRR525 pKa = 11.84NISTVDD531 pKa = 3.24VSHH534 pKa = 6.18QLSYY538 pKa = 11.68SRR540 pKa = 11.84FTLPFKK546 pKa = 10.45IDD548 pKa = 3.53CLKK551 pKa = 9.6DD552 pKa = 3.61QKK554 pKa = 10.49WFASSMTPKK563 pKa = 10.18EE564 pKa = 3.52ISDD567 pKa = 3.5RR568 pKa = 11.84VMEE571 pKa = 4.5ICQYY575 pKa = 8.1PHH577 pKa = 7.02GIIVSDD583 pKa = 3.62YY584 pKa = 10.73SRR586 pKa = 11.84LDD588 pKa = 3.43GHH590 pKa = 6.91VSAADD595 pKa = 3.46KK596 pKa = 10.96AFKK599 pKa = 10.1EE600 pKa = 4.07AVYY603 pKa = 10.29QSWCAHH609 pKa = 7.04AYY611 pKa = 9.63RR612 pKa = 11.84AEE614 pKa = 4.03LSKK617 pKa = 10.86ILAADD622 pKa = 4.42RR623 pKa = 11.84NPKK626 pKa = 10.06GVTAQGLKK634 pKa = 10.06YY635 pKa = 10.64DD636 pKa = 4.2PGYY639 pKa = 10.24SQLSGSPGTTNDD651 pKa = 3.37NNLVTLRR658 pKa = 11.84HH659 pKa = 6.56DD660 pKa = 4.22YY661 pKa = 10.52IALRR665 pKa = 11.84EE666 pKa = 4.15LGNTPKK672 pKa = 10.51QAWQLVQQWVLGASDD687 pKa = 3.5DD688 pKa = 4.34RR689 pKa = 11.84IRR691 pKa = 11.84ANIPGLAQKK700 pKa = 10.74LEE702 pKa = 4.16EE703 pKa = 4.21VATKK707 pKa = 10.45LGHH710 pKa = 5.0QLKK713 pKa = 10.66SIILEE718 pKa = 4.08PLAGCPVPFLGRR730 pKa = 11.84IYY732 pKa = 10.82ASPATHH738 pKa = 6.77NDD740 pKa = 3.02SVQDD744 pKa = 3.87PEE746 pKa = 3.97RR747 pKa = 11.84TLAKK751 pKa = 10.28LHH753 pKa = 6.1LTMSPPTITPLQALYY768 pKa = 10.57NRR770 pKa = 11.84AYY772 pKa = 10.26GYY774 pKa = 11.12YY775 pKa = 8.68VTDD778 pKa = 4.13CKK780 pKa = 10.78TPIIGTWCRR789 pKa = 11.84RR790 pKa = 11.84VIEE793 pKa = 4.02ILEE796 pKa = 4.28TQGMEE801 pKa = 3.83LRR803 pKa = 11.84QATGEE808 pKa = 3.54EE809 pKa = 4.58DD810 pKa = 3.51YY811 pKa = 10.71RR812 pKa = 11.84IKK814 pKa = 10.72SGPYY818 pKa = 7.42PQEE821 pKa = 3.82NEE823 pKa = 3.76EE824 pKa = 4.13LLRR827 pKa = 11.84NLMCQLLDD835 pKa = 3.68LTADD839 pKa = 3.51EE840 pKa = 5.06LDD842 pKa = 4.02SIEE845 pKa = 4.3RR846 pKa = 11.84AINSATTVEE855 pKa = 4.65DD856 pKa = 3.76LPEE859 pKa = 5.43AILDD863 pKa = 3.71NGHH866 pKa = 6.61TIRR869 pKa = 11.84HH870 pKa = 6.34KK871 pKa = 9.85INAAVGHH878 pKa = 7.29DD879 pKa = 3.33ILGPVPSVSSEE890 pKa = 3.78TSEE893 pKa = 3.92PKK895 pKa = 10.3VEE897 pKa = 4.55EE898 pKa = 4.21EE899 pKa = 4.4PKK901 pKa = 10.59CPSVPIISEE910 pKa = 4.33TSASPEE916 pKa = 4.12MIGSDD921 pKa = 3.13ASNPISASPSPPPATATNISHH942 pKa = 6.66PTAGTRR948 pKa = 11.84KK949 pKa = 9.55PSINKK954 pKa = 9.2QSSRR958 pKa = 11.84QLKK961 pKa = 8.82QSSPRR966 pKa = 11.84SQQTKK971 pKa = 8.06HH972 pKa = 5.65QSVNLTPRR980 pKa = 11.84THH982 pKa = 6.1HH983 pKa = 6.38TFNEE987 pKa = 4.52CTRR990 pKa = 11.84KK991 pKa = 10.02DD992 pKa = 2.95ACRR995 pKa = 11.84YY996 pKa = 8.17HH997 pKa = 7.23RR998 pKa = 11.84NGATKK1003 pKa = 9.95PKK1005 pKa = 9.06QTTAPRR1011 pKa = 11.84KK1012 pKa = 9.23
MM1 pKa = 8.03PINIRR6 pKa = 11.84TPVTASPEE14 pKa = 4.12QSSSHH19 pKa = 5.55TPFFCWKK26 pKa = 9.22VFNRR30 pKa = 11.84VQRR33 pKa = 11.84VTPKK37 pKa = 10.19PKK39 pKa = 9.9TSVATAEE46 pKa = 3.92YY47 pKa = 10.52RR48 pKa = 11.84PGKK51 pKa = 9.33VNLMQKK57 pKa = 8.45TRR59 pKa = 11.84DD60 pKa = 3.23RR61 pKa = 11.84FFKK64 pKa = 9.35MWNGNDD70 pKa = 3.11LPKK73 pKa = 10.14RR74 pKa = 11.84SKK76 pKa = 9.79YY77 pKa = 10.37ASNRR81 pKa = 11.84RR82 pKa = 11.84SLPPIDD88 pKa = 3.97FRR90 pKa = 11.84KK91 pKa = 9.3MLHH94 pKa = 6.13HH95 pKa = 6.68HH96 pKa = 6.92LKK98 pKa = 10.34SIPKK102 pKa = 9.37IPMASNHH109 pKa = 4.88THH111 pKa = 6.77PNAASLRR118 pKa = 11.84TSVAVEE124 pKa = 3.7LEE126 pKa = 4.21KK127 pKa = 10.69IVINAGFRR135 pKa = 11.84PYY137 pKa = 10.26SVSMSKK143 pKa = 10.01RR144 pKa = 11.84DD145 pKa = 3.42QYY147 pKa = 11.6DD148 pKa = 3.08GCRR151 pKa = 11.84YY152 pKa = 10.42YY153 pKa = 11.31FMDD156 pKa = 5.05KK157 pKa = 10.99DD158 pKa = 3.43LDD160 pKa = 3.57KK161 pKa = 11.35AFRR164 pKa = 11.84DD165 pKa = 3.6DD166 pKa = 4.31RR167 pKa = 11.84VTEE170 pKa = 3.97EE171 pKa = 4.66HH172 pKa = 6.53VLLMIDD178 pKa = 2.82VDD180 pKa = 4.4YY181 pKa = 11.62YY182 pKa = 11.7CDD184 pKa = 3.41INQYY188 pKa = 10.05LQLGNPIMIYY198 pKa = 9.57TFVPTEE204 pKa = 3.83AGGRR208 pKa = 11.84ALDD211 pKa = 3.93ASYY214 pKa = 10.07TINNNVVSYY223 pKa = 9.36CVKK226 pKa = 10.66GGATYY231 pKa = 10.1HH232 pKa = 6.41HH233 pKa = 6.84EE234 pKa = 3.88LWDD237 pKa = 3.81YY238 pKa = 11.37QGDD241 pKa = 3.96NVVVKK246 pKa = 10.3DD247 pKa = 3.39KK248 pKa = 11.23HH249 pKa = 6.68GNTLVYY255 pKa = 10.53VIEE258 pKa = 3.9QHH260 pKa = 7.18IIEE263 pKa = 4.84EE264 pKa = 4.4DD265 pKa = 3.28PNRR268 pKa = 11.84RR269 pKa = 11.84VIGFYY274 pKa = 10.49PIASYY279 pKa = 9.98PKK281 pKa = 8.19HH282 pKa = 5.41TCPFKK287 pKa = 10.42PEE289 pKa = 4.04SYY291 pKa = 10.76GFGRR295 pKa = 11.84LRR297 pKa = 11.84PSSMGVNCIRR307 pKa = 11.84NVTDD311 pKa = 5.07DD312 pKa = 4.23IISVAVQDD320 pKa = 3.69SANAVTIPQSIYY332 pKa = 10.02DD333 pKa = 3.74ALVVRR338 pKa = 11.84RR339 pKa = 11.84GEE341 pKa = 4.21SKK343 pKa = 10.83NPVIADD349 pKa = 3.17VEE351 pKa = 4.55RR352 pKa = 11.84ILNAEE357 pKa = 4.56HH358 pKa = 7.04IEE360 pKa = 4.04HH361 pKa = 6.47SAIKK365 pKa = 10.59APVLFKK371 pKa = 10.91LLQSSTTSCGVITSSATIGQAKK393 pKa = 9.74NFQTLFPLATEE404 pKa = 4.56DD405 pKa = 3.6GKK407 pKa = 10.26PVGRR411 pKa = 11.84AVAPSLVTDD420 pKa = 4.26PAFVPCKK427 pKa = 10.6SFNNDD432 pKa = 2.38NATIQGRR439 pKa = 11.84VNKK442 pKa = 9.42VRR444 pKa = 11.84NNNQSPPAWKK454 pKa = 8.95TYY456 pKa = 9.95DD457 pKa = 3.33AEE459 pKa = 4.27LVKK462 pKa = 10.6FIVPHH467 pKa = 6.03NKK469 pKa = 9.45NATGVPFDD477 pKa = 3.76YY478 pKa = 10.95EE479 pKa = 4.32RR480 pKa = 11.84VIEE483 pKa = 4.29LQNKK487 pKa = 6.24PAQRR491 pKa = 11.84GRR493 pKa = 11.84SEE495 pKa = 3.89QAKK498 pKa = 8.58ATVSNDD504 pKa = 3.03YY505 pKa = 10.54VNKK508 pKa = 9.83VKK510 pKa = 11.01AFIKK514 pKa = 10.36AEE516 pKa = 4.2AYY518 pKa = 10.44ASVTDD523 pKa = 3.92PRR525 pKa = 11.84NISTVDD531 pKa = 3.24VSHH534 pKa = 6.18QLSYY538 pKa = 11.68SRR540 pKa = 11.84FTLPFKK546 pKa = 10.45IDD548 pKa = 3.53CLKK551 pKa = 9.6DD552 pKa = 3.61QKK554 pKa = 10.49WFASSMTPKK563 pKa = 10.18EE564 pKa = 3.52ISDD567 pKa = 3.5RR568 pKa = 11.84VMEE571 pKa = 4.5ICQYY575 pKa = 8.1PHH577 pKa = 7.02GIIVSDD583 pKa = 3.62YY584 pKa = 10.73SRR586 pKa = 11.84LDD588 pKa = 3.43GHH590 pKa = 6.91VSAADD595 pKa = 3.46KK596 pKa = 10.96AFKK599 pKa = 10.1EE600 pKa = 4.07AVYY603 pKa = 10.29QSWCAHH609 pKa = 7.04AYY611 pKa = 9.63RR612 pKa = 11.84AEE614 pKa = 4.03LSKK617 pKa = 10.86ILAADD622 pKa = 4.42RR623 pKa = 11.84NPKK626 pKa = 10.06GVTAQGLKK634 pKa = 10.06YY635 pKa = 10.64DD636 pKa = 4.2PGYY639 pKa = 10.24SQLSGSPGTTNDD651 pKa = 3.37NNLVTLRR658 pKa = 11.84HH659 pKa = 6.56DD660 pKa = 4.22YY661 pKa = 10.52IALRR665 pKa = 11.84EE666 pKa = 4.15LGNTPKK672 pKa = 10.51QAWQLVQQWVLGASDD687 pKa = 3.5DD688 pKa = 4.34RR689 pKa = 11.84IRR691 pKa = 11.84ANIPGLAQKK700 pKa = 10.74LEE702 pKa = 4.16EE703 pKa = 4.21VATKK707 pKa = 10.45LGHH710 pKa = 5.0QLKK713 pKa = 10.66SIILEE718 pKa = 4.08PLAGCPVPFLGRR730 pKa = 11.84IYY732 pKa = 10.82ASPATHH738 pKa = 6.77NDD740 pKa = 3.02SVQDD744 pKa = 3.87PEE746 pKa = 3.97RR747 pKa = 11.84TLAKK751 pKa = 10.28LHH753 pKa = 6.1LTMSPPTITPLQALYY768 pKa = 10.57NRR770 pKa = 11.84AYY772 pKa = 10.26GYY774 pKa = 11.12YY775 pKa = 8.68VTDD778 pKa = 4.13CKK780 pKa = 10.78TPIIGTWCRR789 pKa = 11.84RR790 pKa = 11.84VIEE793 pKa = 4.02ILEE796 pKa = 4.28TQGMEE801 pKa = 3.83LRR803 pKa = 11.84QATGEE808 pKa = 3.54EE809 pKa = 4.58DD810 pKa = 3.51YY811 pKa = 10.71RR812 pKa = 11.84IKK814 pKa = 10.72SGPYY818 pKa = 7.42PQEE821 pKa = 3.82NEE823 pKa = 3.76EE824 pKa = 4.13LLRR827 pKa = 11.84NLMCQLLDD835 pKa = 3.68LTADD839 pKa = 3.51EE840 pKa = 5.06LDD842 pKa = 4.02SIEE845 pKa = 4.3RR846 pKa = 11.84AINSATTVEE855 pKa = 4.65DD856 pKa = 3.76LPEE859 pKa = 5.43AILDD863 pKa = 3.71NGHH866 pKa = 6.61TIRR869 pKa = 11.84HH870 pKa = 6.34KK871 pKa = 9.85INAAVGHH878 pKa = 7.29DD879 pKa = 3.33ILGPVPSVSSEE890 pKa = 3.78TSEE893 pKa = 3.92PKK895 pKa = 10.3VEE897 pKa = 4.55EE898 pKa = 4.21EE899 pKa = 4.4PKK901 pKa = 10.59CPSVPIISEE910 pKa = 4.33TSASPEE916 pKa = 4.12MIGSDD921 pKa = 3.13ASNPISASPSPPPATATNISHH942 pKa = 6.66PTAGTRR948 pKa = 11.84KK949 pKa = 9.55PSINKK954 pKa = 9.2QSSRR958 pKa = 11.84QLKK961 pKa = 8.82QSSPRR966 pKa = 11.84SQQTKK971 pKa = 8.06HH972 pKa = 5.65QSVNLTPRR980 pKa = 11.84THH982 pKa = 6.1HH983 pKa = 6.38TFNEE987 pKa = 4.52CTRR990 pKa = 11.84KK991 pKa = 10.02DD992 pKa = 2.95ACRR995 pKa = 11.84YY996 pKa = 8.17HH997 pKa = 7.23RR998 pKa = 11.84NGATKK1003 pKa = 9.95PKK1005 pKa = 9.06QTTAPRR1011 pKa = 11.84KK1012 pKa = 9.23
Molecular weight: 112.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG99|A0A1L3KG99_9VIRU Capsid protein OS=Sanxia water strider virus 17 OX=1923401 PE=3 SV=1
MM1 pKa = 7.92APQNQNKK8 pKa = 7.12QQPRR12 pKa = 11.84ASKK15 pKa = 10.23RR16 pKa = 11.84SQRR19 pKa = 11.84TNMNQQRR26 pKa = 11.84TVASVPRR33 pKa = 11.84QGLRR37 pKa = 11.84SKK39 pKa = 10.14QQTYY43 pKa = 7.26MTGQPKK49 pKa = 9.41FQHH52 pKa = 6.56KK53 pKa = 8.99GDD55 pKa = 3.66KK56 pKa = 10.75VIVSNSEE63 pKa = 3.41ILVEE67 pKa = 4.33VNGTVASGVIPAGGAIRR84 pKa = 11.84VFRR87 pKa = 11.84FEE89 pKa = 4.89NVATGNNMNTTRR101 pKa = 11.84WLTGMAKK108 pKa = 10.3LYY110 pKa = 11.09DD111 pKa = 3.58KK112 pKa = 11.14FRR114 pKa = 11.84VNKK117 pKa = 9.73MNMRR121 pKa = 11.84WVTSVPFTYY130 pKa = 10.23GGQVALRR137 pKa = 11.84FDD139 pKa = 4.19SDD141 pKa = 3.82PSKK144 pKa = 8.39TTADD148 pKa = 3.37ASLLAVSGDD157 pKa = 3.33MRR159 pKa = 11.84ALATAVYY166 pKa = 9.69NSASNRR172 pKa = 11.84VKK174 pKa = 10.6KK175 pKa = 10.48DD176 pKa = 3.02QLNRR180 pKa = 11.84LPQYY184 pKa = 7.97EE185 pKa = 4.54TFPSATDD192 pKa = 3.21TGVATVGSINLAYY205 pKa = 10.51SAITPPAGTTGTINLGYY222 pKa = 10.2VWVDD226 pKa = 3.06YY227 pKa = 10.59EE228 pKa = 4.76VEE230 pKa = 4.31FFNPSNAIAAA240 pKa = 4.21
MM1 pKa = 7.92APQNQNKK8 pKa = 7.12QQPRR12 pKa = 11.84ASKK15 pKa = 10.23RR16 pKa = 11.84SQRR19 pKa = 11.84TNMNQQRR26 pKa = 11.84TVASVPRR33 pKa = 11.84QGLRR37 pKa = 11.84SKK39 pKa = 10.14QQTYY43 pKa = 7.26MTGQPKK49 pKa = 9.41FQHH52 pKa = 6.56KK53 pKa = 8.99GDD55 pKa = 3.66KK56 pKa = 10.75VIVSNSEE63 pKa = 3.41ILVEE67 pKa = 4.33VNGTVASGVIPAGGAIRR84 pKa = 11.84VFRR87 pKa = 11.84FEE89 pKa = 4.89NVATGNNMNTTRR101 pKa = 11.84WLTGMAKK108 pKa = 10.3LYY110 pKa = 11.09DD111 pKa = 3.58KK112 pKa = 11.14FRR114 pKa = 11.84VNKK117 pKa = 9.73MNMRR121 pKa = 11.84WVTSVPFTYY130 pKa = 10.23GGQVALRR137 pKa = 11.84FDD139 pKa = 4.19SDD141 pKa = 3.82PSKK144 pKa = 8.39TTADD148 pKa = 3.37ASLLAVSGDD157 pKa = 3.33MRR159 pKa = 11.84ALATAVYY166 pKa = 9.69NSASNRR172 pKa = 11.84VKK174 pKa = 10.6KK175 pKa = 10.48DD176 pKa = 3.02QLNRR180 pKa = 11.84LPQYY184 pKa = 7.97EE185 pKa = 4.54TFPSATDD192 pKa = 3.21TGVATVGSINLAYY205 pKa = 10.51SAITPPAGTTGTINLGYY222 pKa = 10.2VWVDD226 pKa = 3.06YY227 pKa = 10.59EE228 pKa = 4.76VEE230 pKa = 4.31FFNPSNAIAAA240 pKa = 4.21
Molecular weight: 26.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1252 |
240 |
1012 |
626.0 |
69.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.227 ± 0.843 | 1.438 ± 0.684 |
5.112 ± 0.648 | 4.553 ± 0.976 |
2.875 ± 0.416 | 5.112 ± 0.938 |
2.556 ± 1.017 | 5.671 ± 1.112 |
5.911 ± 0.433 | 6.23 ± 0.585 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.917 ± 0.674 | 5.911 ± 0.954 |
6.869 ± 0.889 | 4.633 ± 0.769 |
5.911 ± 0.161 | 7.907 ± 0.194 |
7.109 ± 0.979 | 7.348 ± 0.865 |
0.958 ± 0.139 | 3.754 ± 0.2 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
