
Phaeobacter sp. CECT 5382
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Phaeobacter; unclassified Phaeobacter
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
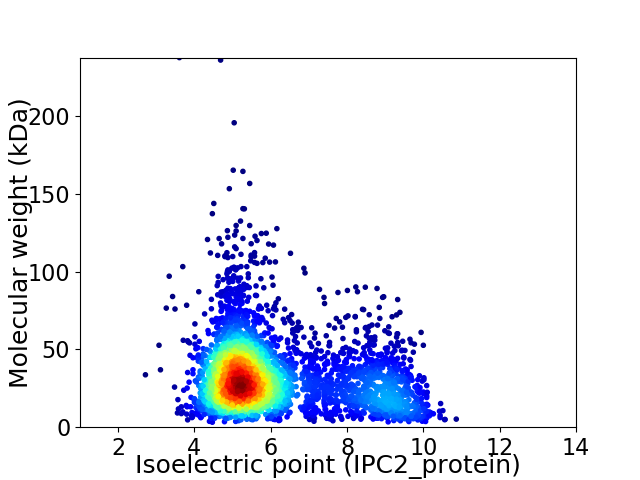
Virtual 2D-PAGE plot for 3864 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
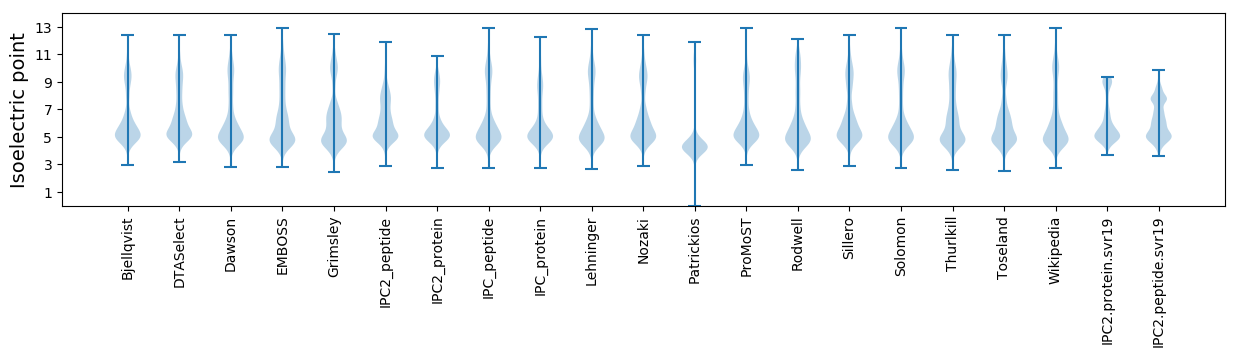
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P1GXQ1|A0A0P1GXQ1_9RHOB p-hydroxybenzoate hydroxylase OS=Phaeobacter sp. CECT 5382 OX=1712645 GN=pobA PE=4 SV=1
MM1 pKa = 7.15ATATEE6 pKa = 4.17LDD8 pKa = 3.69INTSATATQMANEE21 pKa = 4.13IFGDD25 pKa = 3.75GTTVVSASYY34 pKa = 9.81TGDD37 pKa = 3.28VDD39 pKa = 4.2SSGIYY44 pKa = 9.35TGADD48 pKa = 3.03TTVAGVAPADD58 pKa = 3.49NGVILSTGDD67 pKa = 3.39VVDD70 pKa = 4.49FTNNSGTTNTNTSGNTSTNTSGVNGDD96 pKa = 3.45SDD98 pKa = 4.12FNAIAGSATYY108 pKa = 10.31DD109 pKa = 3.33AAFLEE114 pKa = 4.74IDD116 pKa = 5.04FIPDD120 pKa = 3.32GDD122 pKa = 4.26VLTLDD127 pKa = 4.47FVLSSEE133 pKa = 4.73EE134 pKa = 4.0YY135 pKa = 10.18PEE137 pKa = 4.54FASSQYY143 pKa = 10.97NDD145 pKa = 3.52VVGVWVNGVQAQVTIGDD162 pKa = 4.2GSASIGNINQSDD174 pKa = 3.9TQNIYY179 pKa = 10.83VDD181 pKa = 3.6NTSDD185 pKa = 3.72QFNTEE190 pKa = 3.6MDD192 pKa = 3.68GFTITLTFTAPVNAGAVNSIKK213 pKa = 11.0VGVADD218 pKa = 4.49VADD221 pKa = 3.75SSYY224 pKa = 10.74DD225 pKa = 3.65TNLLIAGGSVQSAFVAQDD243 pKa = 3.6DD244 pKa = 4.62SLTMPLAATRR254 pKa = 11.84MLDD257 pKa = 4.11VLDD260 pKa = 4.67NDD262 pKa = 3.89AATAGGTLTITHH274 pKa = 6.72INDD277 pKa = 3.24VAVTAGDD284 pKa = 3.96TVTLSSGQQITLNADD299 pKa = 3.09GTLEE303 pKa = 4.11VVADD307 pKa = 4.19GDD309 pKa = 4.14AEE311 pKa = 4.35TVYY314 pKa = 10.79FNYY317 pKa = 9.88TVEE320 pKa = 4.15NDD322 pKa = 4.01LGDD325 pKa = 3.75SDD327 pKa = 4.13TALVEE332 pKa = 3.91ITQAQPCFLRR342 pKa = 11.84GTLIVTEE349 pKa = 5.04DD350 pKa = 3.84GPLPIEE356 pKa = 4.4DD357 pKa = 5.06LRR359 pKa = 11.84PGMLVEE365 pKa = 4.75TYY367 pKa = 11.23DD368 pKa = 4.7HH369 pKa = 7.06GLQPLRR375 pKa = 11.84WIGHH379 pKa = 4.31SHH381 pKa = 4.35VTVSEE386 pKa = 4.15TTAPIRR392 pKa = 11.84FKK394 pKa = 10.62KK395 pKa = 9.1GQYY398 pKa = 10.11GCEE401 pKa = 3.68QDD403 pKa = 4.52LWVSPNHH410 pKa = 6.13RR411 pKa = 11.84MLVTGQFAEE420 pKa = 4.27LLFGEE425 pKa = 4.65SEE427 pKa = 4.46VLAKK431 pKa = 10.8AKK433 pKa = 10.7DD434 pKa = 3.77LVNDD438 pKa = 3.5SSIRR442 pKa = 11.84PDD444 pKa = 3.08HH445 pKa = 6.52SFEE448 pKa = 4.09TVEE451 pKa = 4.0YY452 pKa = 9.97FHH454 pKa = 7.97LLFDD458 pKa = 3.32QHH460 pKa = 6.9EE461 pKa = 4.51ILWSNGALSEE471 pKa = 4.51SYY473 pKa = 10.73HH474 pKa = 6.54PGPEE478 pKa = 3.89TTRR481 pKa = 11.84GFDD484 pKa = 3.56AEE486 pKa = 4.39TQAEE490 pKa = 4.47LFALFPEE497 pKa = 5.14LCPMTGTGYY506 pKa = 10.59GAAARR511 pKa = 11.84LSLRR515 pKa = 11.84HH516 pKa = 6.02FEE518 pKa = 4.27TVLLDD523 pKa = 3.63TYY525 pKa = 10.88MSS527 pKa = 3.64
MM1 pKa = 7.15ATATEE6 pKa = 4.17LDD8 pKa = 3.69INTSATATQMANEE21 pKa = 4.13IFGDD25 pKa = 3.75GTTVVSASYY34 pKa = 9.81TGDD37 pKa = 3.28VDD39 pKa = 4.2SSGIYY44 pKa = 9.35TGADD48 pKa = 3.03TTVAGVAPADD58 pKa = 3.49NGVILSTGDD67 pKa = 3.39VVDD70 pKa = 4.49FTNNSGTTNTNTSGNTSTNTSGVNGDD96 pKa = 3.45SDD98 pKa = 4.12FNAIAGSATYY108 pKa = 10.31DD109 pKa = 3.33AAFLEE114 pKa = 4.74IDD116 pKa = 5.04FIPDD120 pKa = 3.32GDD122 pKa = 4.26VLTLDD127 pKa = 4.47FVLSSEE133 pKa = 4.73EE134 pKa = 4.0YY135 pKa = 10.18PEE137 pKa = 4.54FASSQYY143 pKa = 10.97NDD145 pKa = 3.52VVGVWVNGVQAQVTIGDD162 pKa = 4.2GSASIGNINQSDD174 pKa = 3.9TQNIYY179 pKa = 10.83VDD181 pKa = 3.6NTSDD185 pKa = 3.72QFNTEE190 pKa = 3.6MDD192 pKa = 3.68GFTITLTFTAPVNAGAVNSIKK213 pKa = 11.0VGVADD218 pKa = 4.49VADD221 pKa = 3.75SSYY224 pKa = 10.74DD225 pKa = 3.65TNLLIAGGSVQSAFVAQDD243 pKa = 3.6DD244 pKa = 4.62SLTMPLAATRR254 pKa = 11.84MLDD257 pKa = 4.11VLDD260 pKa = 4.67NDD262 pKa = 3.89AATAGGTLTITHH274 pKa = 6.72INDD277 pKa = 3.24VAVTAGDD284 pKa = 3.96TVTLSSGQQITLNADD299 pKa = 3.09GTLEE303 pKa = 4.11VVADD307 pKa = 4.19GDD309 pKa = 4.14AEE311 pKa = 4.35TVYY314 pKa = 10.79FNYY317 pKa = 9.88TVEE320 pKa = 4.15NDD322 pKa = 4.01LGDD325 pKa = 3.75SDD327 pKa = 4.13TALVEE332 pKa = 3.91ITQAQPCFLRR342 pKa = 11.84GTLIVTEE349 pKa = 5.04DD350 pKa = 3.84GPLPIEE356 pKa = 4.4DD357 pKa = 5.06LRR359 pKa = 11.84PGMLVEE365 pKa = 4.75TYY367 pKa = 11.23DD368 pKa = 4.7HH369 pKa = 7.06GLQPLRR375 pKa = 11.84WIGHH379 pKa = 4.31SHH381 pKa = 4.35VTVSEE386 pKa = 4.15TTAPIRR392 pKa = 11.84FKK394 pKa = 10.62KK395 pKa = 9.1GQYY398 pKa = 10.11GCEE401 pKa = 3.68QDD403 pKa = 4.52LWVSPNHH410 pKa = 6.13RR411 pKa = 11.84MLVTGQFAEE420 pKa = 4.27LLFGEE425 pKa = 4.65SEE427 pKa = 4.46VLAKK431 pKa = 10.8AKK433 pKa = 10.7DD434 pKa = 3.77LVNDD438 pKa = 3.5SSIRR442 pKa = 11.84PDD444 pKa = 3.08HH445 pKa = 6.52SFEE448 pKa = 4.09TVEE451 pKa = 4.0YY452 pKa = 9.97FHH454 pKa = 7.97LLFDD458 pKa = 3.32QHH460 pKa = 6.9EE461 pKa = 4.51ILWSNGALSEE471 pKa = 4.51SYY473 pKa = 10.73HH474 pKa = 6.54PGPEE478 pKa = 3.89TTRR481 pKa = 11.84GFDD484 pKa = 3.56AEE486 pKa = 4.39TQAEE490 pKa = 4.47LFALFPEE497 pKa = 5.14LCPMTGTGYY506 pKa = 10.59GAAARR511 pKa = 11.84LSLRR515 pKa = 11.84HH516 pKa = 6.02FEE518 pKa = 4.27TVLLDD523 pKa = 3.63TYY525 pKa = 10.88MSS527 pKa = 3.64
Molecular weight: 55.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P1H4K5|A0A0P1H4K5_9RHOB Uncharacterized protein OS=Phaeobacter sp. CECT 5382 OX=1712645 GN=PH5382_03865 PE=4 SV=1
MM1 pKa = 6.84TACPTFAGPALFSYY15 pKa = 9.41GFRR18 pKa = 11.84PFFAAACIFGFLVVPLWVLIWRR40 pKa = 11.84GSLEE44 pKa = 4.22YY45 pKa = 9.96QGHH48 pKa = 6.36FGAVDD53 pKa = 3.07WHH55 pKa = 6.08IHH57 pKa = 4.59EE58 pKa = 5.21MIFGYY63 pKa = 10.66GSAVLAGFLFTAVPNWTARR82 pKa = 11.84MPVRR86 pKa = 11.84GLPLAVLFAVWILGRR101 pKa = 11.84LALWGGLVAPGLALVVEE118 pKa = 5.07CSFLLLVAAMIARR131 pKa = 11.84EE132 pKa = 4.06IIAGKK137 pKa = 8.27NWRR140 pKa = 11.84NLKK143 pKa = 10.33VLVPVLLLGAGNILFHH159 pKa = 8.03LEE161 pKa = 4.23VILSGASDD169 pKa = 3.36YY170 pKa = 11.34GRR172 pKa = 11.84RR173 pKa = 11.84FGIALLVFLIMLIGGRR189 pKa = 11.84IIPSFTRR196 pKa = 11.84NWLAKK201 pKa = 10.08EE202 pKa = 4.07RR203 pKa = 11.84GQDD206 pKa = 3.42GPMPIPFSRR215 pKa = 11.84FDD217 pKa = 3.57AVALAFGVAALVGWTLAPEE236 pKa = 4.44SVVSAVLLLLAGVLHH251 pKa = 7.33IARR254 pKa = 11.84LTRR257 pKa = 11.84WQGQAVLRR265 pKa = 11.84SPLLVMLHH273 pKa = 5.56VAYY276 pKa = 9.83IFIPLGLMVSGFAALGLVEE295 pKa = 4.49QVAAVHH301 pKa = 6.17LLGIGAIGGMTVAVMMRR318 pKa = 11.84ATLGHH323 pKa = 6.2TGRR326 pKa = 11.84ALVAGPVLTLGFGLLLAAAVARR348 pKa = 11.84IGSGILADD356 pKa = 4.4LGLDD360 pKa = 3.68GVLLSAGFWTVSFAVLCLRR379 pKa = 11.84MLPWLVLPKK388 pKa = 10.22AARR391 pKa = 11.84RR392 pKa = 11.84KK393 pKa = 9.47PNPAPQQAVNPSPKK407 pKa = 9.93VAGG410 pKa = 3.68
MM1 pKa = 6.84TACPTFAGPALFSYY15 pKa = 9.41GFRR18 pKa = 11.84PFFAAACIFGFLVVPLWVLIWRR40 pKa = 11.84GSLEE44 pKa = 4.22YY45 pKa = 9.96QGHH48 pKa = 6.36FGAVDD53 pKa = 3.07WHH55 pKa = 6.08IHH57 pKa = 4.59EE58 pKa = 5.21MIFGYY63 pKa = 10.66GSAVLAGFLFTAVPNWTARR82 pKa = 11.84MPVRR86 pKa = 11.84GLPLAVLFAVWILGRR101 pKa = 11.84LALWGGLVAPGLALVVEE118 pKa = 5.07CSFLLLVAAMIARR131 pKa = 11.84EE132 pKa = 4.06IIAGKK137 pKa = 8.27NWRR140 pKa = 11.84NLKK143 pKa = 10.33VLVPVLLLGAGNILFHH159 pKa = 8.03LEE161 pKa = 4.23VILSGASDD169 pKa = 3.36YY170 pKa = 11.34GRR172 pKa = 11.84RR173 pKa = 11.84FGIALLVFLIMLIGGRR189 pKa = 11.84IIPSFTRR196 pKa = 11.84NWLAKK201 pKa = 10.08EE202 pKa = 4.07RR203 pKa = 11.84GQDD206 pKa = 3.42GPMPIPFSRR215 pKa = 11.84FDD217 pKa = 3.57AVALAFGVAALVGWTLAPEE236 pKa = 4.44SVVSAVLLLLAGVLHH251 pKa = 7.33IARR254 pKa = 11.84LTRR257 pKa = 11.84WQGQAVLRR265 pKa = 11.84SPLLVMLHH273 pKa = 5.56VAYY276 pKa = 9.83IFIPLGLMVSGFAALGLVEE295 pKa = 4.49QVAAVHH301 pKa = 6.17LLGIGAIGGMTVAVMMRR318 pKa = 11.84ATLGHH323 pKa = 6.2TGRR326 pKa = 11.84ALVAGPVLTLGFGLLLAAAVARR348 pKa = 11.84IGSGILADD356 pKa = 4.4LGLDD360 pKa = 3.68GVLLSAGFWTVSFAVLCLRR379 pKa = 11.84MLPWLVLPKK388 pKa = 10.22AARR391 pKa = 11.84RR392 pKa = 11.84KK393 pKa = 9.47PNPAPQQAVNPSPKK407 pKa = 9.93VAGG410 pKa = 3.68
Molecular weight: 43.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1199402 |
30 |
2393 |
310.4 |
33.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.911 ± 0.055 | 0.975 ± 0.013 |
5.848 ± 0.032 | 5.946 ± 0.038 |
3.765 ± 0.027 | 8.44 ± 0.041 |
2.115 ± 0.02 | 5.106 ± 0.028 |
3.523 ± 0.032 | 10.321 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.82 ± 0.018 | 2.745 ± 0.02 |
4.902 ± 0.029 | 3.802 ± 0.025 |
6.23 ± 0.037 | 5.628 ± 0.034 |
5.356 ± 0.031 | 6.94 ± 0.031 |
1.375 ± 0.017 | 2.254 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
