
Pectobacterium phage Peat1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Corkvirinae; Phimunavirus; Pectobacterium virus Peat1
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
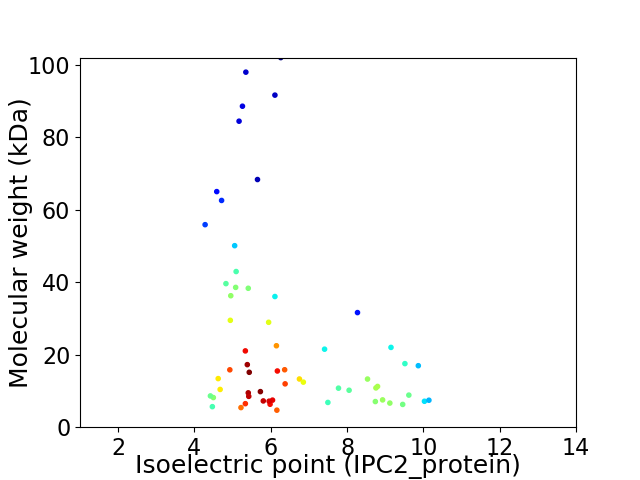
Virtual 2D-PAGE plot for 61 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0H3YI42|A0A0H3YI42_9CAUD Type VII integral membrane protein OS=Pectobacterium phage Peat1 OX=1654601 PE=4 SV=1
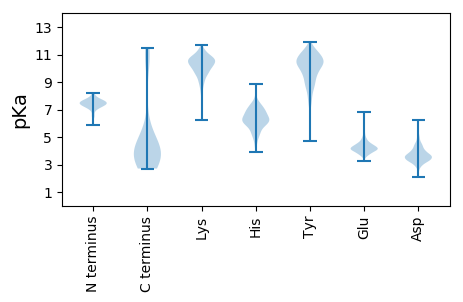
MM1 pKa = 7.42YY2 pKa = 9.95SVQIAVSDD10 pKa = 3.84GTLTRR15 pKa = 11.84IALSIEE21 pKa = 4.16YY22 pKa = 10.07FEE24 pKa = 6.26KK25 pKa = 10.91DD26 pKa = 4.32DD27 pKa = 3.3ITLYY31 pKa = 10.93RR32 pKa = 11.84NLEE35 pKa = 4.1LTPLVLGTDD44 pKa = 3.81WQWDD48 pKa = 3.51GDD50 pKa = 3.9THH52 pKa = 7.88INLLTGIPVPVGSYY66 pKa = 8.03ITVRR70 pKa = 11.84RR71 pKa = 11.84NTDD74 pKa = 2.11IDD76 pKa = 3.69RR77 pKa = 11.84AFNIYY82 pKa = 10.62DD83 pKa = 3.47GGAAFNRR90 pKa = 11.84EE91 pKa = 3.99TLDD94 pKa = 3.47EE95 pKa = 4.33NFKK98 pKa = 11.24QMIYY102 pKa = 10.2LAQEE106 pKa = 3.97FTEE109 pKa = 4.83GNGLTGLYY117 pKa = 10.06FPLDD121 pKa = 3.21MHH123 pKa = 6.89GFQIKK128 pKa = 10.46NLGEE132 pKa = 4.05PTDD135 pKa = 4.43PGDD138 pKa = 4.01AVTKK142 pKa = 10.37QYY144 pKa = 11.57VDD146 pKa = 3.51TANTAQNANFNASQQAQDD164 pKa = 3.44QAVAASQAVQDD175 pKa = 3.98NRR177 pKa = 11.84LASLEE182 pKa = 4.13NTFVQATSSYY192 pKa = 8.88PWYY195 pKa = 9.16TVSTSTTDD203 pKa = 2.82TFTPGFNFTKK213 pKa = 10.29AAVYY217 pKa = 10.47INGVCQTPDD226 pKa = 2.85YY227 pKa = 11.04SYY229 pKa = 11.21IVVANQILLADD240 pKa = 4.48PVPLGTMVFARR251 pKa = 11.84LGEE254 pKa = 4.81DD255 pKa = 3.1IQNDD259 pKa = 3.52DD260 pKa = 4.86DD261 pKa = 4.73FATTAQLSAVQANLQDD277 pKa = 4.64EE278 pKa = 4.5IDD280 pKa = 3.8VTNTEE285 pKa = 4.19VSNKK289 pKa = 9.57ASKK292 pKa = 10.13GANSDD297 pKa = 3.1ISSLSGLTTPLSAAQGGTGNNTGNAATATVLATHH331 pKa = 6.42RR332 pKa = 11.84TIQTNLASTSAASFNGSANITPGVTGNLPVTNGGTGAGSAPSARR376 pKa = 11.84ANLGAAMNGINNDD389 pKa = 3.02ISNLAALTGGITGLTTGTAAASGIVGEE416 pKa = 4.5VASAASSSATNLVSGSVINIISLSLPAGDD445 pKa = 4.53WEE447 pKa = 4.82LEE449 pKa = 4.2SAFQIINTGNVTALAFGVSTTTGVLPTPWYY479 pKa = 9.9DD480 pKa = 3.28VYY482 pKa = 11.5SITTTIGSGTSNRR495 pKa = 11.84QGMARR500 pKa = 11.84RR501 pKa = 11.84VLLSTTTTVYY511 pKa = 10.73LVAQATFTGTATANGYY527 pKa = 8.94VRR529 pKa = 11.84ARR531 pKa = 11.84RR532 pKa = 11.84VRR534 pKa = 3.3
MM1 pKa = 7.42YY2 pKa = 9.95SVQIAVSDD10 pKa = 3.84GTLTRR15 pKa = 11.84IALSIEE21 pKa = 4.16YY22 pKa = 10.07FEE24 pKa = 6.26KK25 pKa = 10.91DD26 pKa = 4.32DD27 pKa = 3.3ITLYY31 pKa = 10.93RR32 pKa = 11.84NLEE35 pKa = 4.1LTPLVLGTDD44 pKa = 3.81WQWDD48 pKa = 3.51GDD50 pKa = 3.9THH52 pKa = 7.88INLLTGIPVPVGSYY66 pKa = 8.03ITVRR70 pKa = 11.84RR71 pKa = 11.84NTDD74 pKa = 2.11IDD76 pKa = 3.69RR77 pKa = 11.84AFNIYY82 pKa = 10.62DD83 pKa = 3.47GGAAFNRR90 pKa = 11.84EE91 pKa = 3.99TLDD94 pKa = 3.47EE95 pKa = 4.33NFKK98 pKa = 11.24QMIYY102 pKa = 10.2LAQEE106 pKa = 3.97FTEE109 pKa = 4.83GNGLTGLYY117 pKa = 10.06FPLDD121 pKa = 3.21MHH123 pKa = 6.89GFQIKK128 pKa = 10.46NLGEE132 pKa = 4.05PTDD135 pKa = 4.43PGDD138 pKa = 4.01AVTKK142 pKa = 10.37QYY144 pKa = 11.57VDD146 pKa = 3.51TANTAQNANFNASQQAQDD164 pKa = 3.44QAVAASQAVQDD175 pKa = 3.98NRR177 pKa = 11.84LASLEE182 pKa = 4.13NTFVQATSSYY192 pKa = 8.88PWYY195 pKa = 9.16TVSTSTTDD203 pKa = 2.82TFTPGFNFTKK213 pKa = 10.29AAVYY217 pKa = 10.47INGVCQTPDD226 pKa = 2.85YY227 pKa = 11.04SYY229 pKa = 11.21IVVANQILLADD240 pKa = 4.48PVPLGTMVFARR251 pKa = 11.84LGEE254 pKa = 4.81DD255 pKa = 3.1IQNDD259 pKa = 3.52DD260 pKa = 4.86DD261 pKa = 4.73FATTAQLSAVQANLQDD277 pKa = 4.64EE278 pKa = 4.5IDD280 pKa = 3.8VTNTEE285 pKa = 4.19VSNKK289 pKa = 9.57ASKK292 pKa = 10.13GANSDD297 pKa = 3.1ISSLSGLTTPLSAAQGGTGNNTGNAATATVLATHH331 pKa = 6.42RR332 pKa = 11.84TIQTNLASTSAASFNGSANITPGVTGNLPVTNGGTGAGSAPSARR376 pKa = 11.84ANLGAAMNGINNDD389 pKa = 3.02ISNLAALTGGITGLTTGTAAASGIVGEE416 pKa = 4.5VASAASSSATNLVSGSVINIISLSLPAGDD445 pKa = 4.53WEE447 pKa = 4.82LEE449 pKa = 4.2SAFQIINTGNVTALAFGVSTTTGVLPTPWYY479 pKa = 9.9DD480 pKa = 3.28VYY482 pKa = 11.5SITTTIGSGTSNRR495 pKa = 11.84QGMARR500 pKa = 11.84RR501 pKa = 11.84VLLSTTTTVYY511 pKa = 10.73LVAQATFTGTATANGYY527 pKa = 8.94VRR529 pKa = 11.84ARR531 pKa = 11.84RR532 pKa = 11.84VRR534 pKa = 3.3
Molecular weight: 55.87 kDa
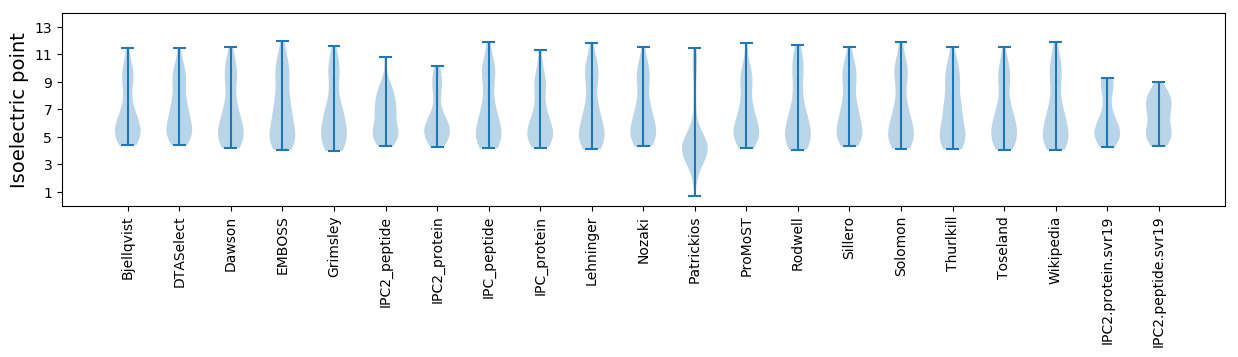
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0H3YI90|A0A0H3YI90_9CAUD Uncharacterized protein OS=Pectobacterium phage Peat1 OX=1654601 PE=4 SV=1
MM1 pKa = 7.63SDD3 pKa = 2.58WYY5 pKa = 10.54YY6 pKa = 11.2DD7 pKa = 3.51KK8 pKa = 11.26EE9 pKa = 4.56SGVVYY14 pKa = 10.67RR15 pKa = 11.84NGRR18 pKa = 11.84PMTLLGKK25 pKa = 10.04DD26 pKa = 3.42GYY28 pKa = 11.79VRR30 pKa = 11.84MRR32 pKa = 11.84YY33 pKa = 9.19KK34 pKa = 10.81GRR36 pKa = 11.84VEE38 pKa = 3.8RR39 pKa = 11.84AHH41 pKa = 6.28RR42 pKa = 11.84LAYY45 pKa = 9.47ILQGLPLPKK54 pKa = 10.04QVDD57 pKa = 4.17HH58 pKa = 7.16INGNRR63 pKa = 11.84ADD65 pKa = 3.9NRR67 pKa = 11.84WVNLRR72 pKa = 11.84PATNTQNQYY81 pKa = 9.2NRR83 pKa = 11.84RR84 pKa = 11.84PASKK88 pKa = 10.26LGHH91 pKa = 6.28KK92 pKa = 9.88KK93 pKa = 9.55GAYY96 pKa = 8.25MNIGGRR102 pKa = 11.84TWYY105 pKa = 10.78SLLRR109 pKa = 11.84YY110 pKa = 8.55NGKK113 pKa = 10.12RR114 pKa = 11.84IYY116 pKa = 10.75LGTFKK121 pKa = 10.75TEE123 pKa = 4.3DD124 pKa = 3.78EE125 pKa = 3.93AHH127 pKa = 6.27AAYY130 pKa = 9.37TEE132 pKa = 4.11ASKK135 pKa = 10.61KK136 pKa = 7.97YY137 pKa = 9.36HH138 pKa = 6.29GNFSRR143 pKa = 11.84TEE145 pKa = 3.65
MM1 pKa = 7.63SDD3 pKa = 2.58WYY5 pKa = 10.54YY6 pKa = 11.2DD7 pKa = 3.51KK8 pKa = 11.26EE9 pKa = 4.56SGVVYY14 pKa = 10.67RR15 pKa = 11.84NGRR18 pKa = 11.84PMTLLGKK25 pKa = 10.04DD26 pKa = 3.42GYY28 pKa = 11.79VRR30 pKa = 11.84MRR32 pKa = 11.84YY33 pKa = 9.19KK34 pKa = 10.81GRR36 pKa = 11.84VEE38 pKa = 3.8RR39 pKa = 11.84AHH41 pKa = 6.28RR42 pKa = 11.84LAYY45 pKa = 9.47ILQGLPLPKK54 pKa = 10.04QVDD57 pKa = 4.17HH58 pKa = 7.16INGNRR63 pKa = 11.84ADD65 pKa = 3.9NRR67 pKa = 11.84WVNLRR72 pKa = 11.84PATNTQNQYY81 pKa = 9.2NRR83 pKa = 11.84RR84 pKa = 11.84PASKK88 pKa = 10.26LGHH91 pKa = 6.28KK92 pKa = 9.88KK93 pKa = 9.55GAYY96 pKa = 8.25MNIGGRR102 pKa = 11.84TWYY105 pKa = 10.78SLLRR109 pKa = 11.84YY110 pKa = 8.55NGKK113 pKa = 10.12RR114 pKa = 11.84IYY116 pKa = 10.75LGTFKK121 pKa = 10.75TEE123 pKa = 4.3DD124 pKa = 3.78EE125 pKa = 3.93AHH127 pKa = 6.27AAYY130 pKa = 9.37TEE132 pKa = 4.11ASKK135 pKa = 10.61KK136 pKa = 7.97YY137 pKa = 9.36HH138 pKa = 6.29GNFSRR143 pKa = 11.84TEE145 pKa = 3.65
Molecular weight: 16.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14173 |
40 |
939 |
232.3 |
25.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.829 ± 0.48 | 1.192 ± 0.187 |
5.969 ± 0.206 | 5.172 ± 0.33 |
2.921 ± 0.143 | 7.571 ± 0.268 |
1.849 ± 0.2 | 4.572 ± 0.22 |
4.502 ± 0.249 | 8.707 ± 0.249 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.568 ± 0.186 | 4.389 ± 0.299 |
3.965 ± 0.21 | 4.89 ± 0.435 |
5.475 ± 0.279 | 6.632 ± 0.266 |
6.738 ± 0.419 | 7.712 ± 0.259 |
1.383 ± 0.145 | 3.93 ± 0.26 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
