
Myroides odoratus DSM 2801
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Myroides; Myroides odoratus
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
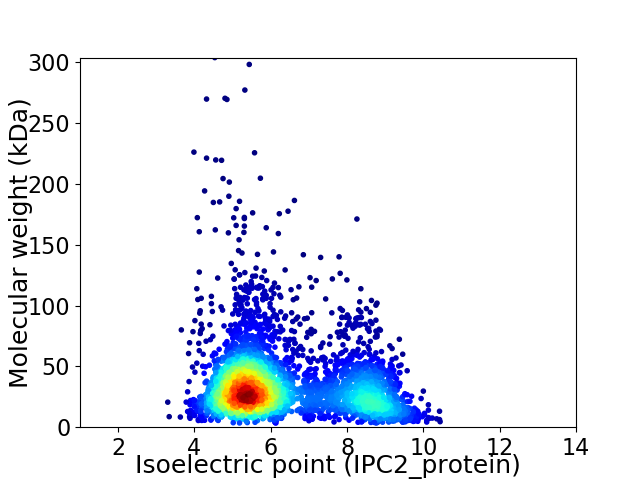
Virtual 2D-PAGE plot for 3687 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H1Z978|H1Z978_MYROD Transporter hydrophobe/amphiphile efflux-1 (HAE1) family OS=Myroides odoratus DSM 2801 OX=929704 GN=Myrod_2783 PE=3 SV=1
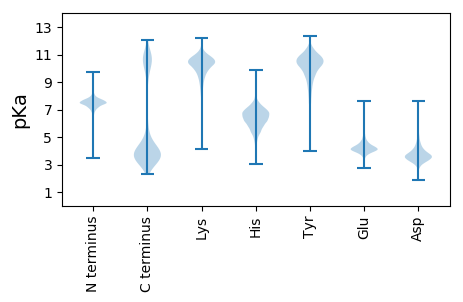
MM1 pKa = 7.49TNNFEE6 pKa = 4.36EE7 pKa = 4.86IINRR11 pKa = 11.84TEE13 pKa = 3.82VQEE16 pKa = 4.11VLNQYY21 pKa = 8.08ITEE24 pKa = 4.22NGGNIHH30 pKa = 6.63YY31 pKa = 10.48DD32 pKa = 3.3GGSFTYY38 pKa = 10.63LDD40 pKa = 3.86EE41 pKa = 4.87NGNTVTMDD49 pKa = 3.3MTTIIQGSEE58 pKa = 4.0TVTTLVKK65 pKa = 10.84DD66 pKa = 3.51PLNNGKK72 pKa = 10.23YY73 pKa = 9.93IYY75 pKa = 9.73TNEE78 pKa = 4.07AGVEE82 pKa = 4.18VVIDD86 pKa = 3.51VTTEE90 pKa = 3.51VTNNFEE96 pKa = 4.27EE97 pKa = 4.81IINRR101 pKa = 11.84TEE103 pKa = 3.82VQEE106 pKa = 4.11VLNQYY111 pKa = 8.08ITEE114 pKa = 4.22NGGNIHH120 pKa = 6.63YY121 pKa = 10.48DD122 pKa = 3.3GGSFTYY128 pKa = 10.63LDD130 pKa = 3.86EE131 pKa = 4.87NGNTVTMDD139 pKa = 3.02MTTIVQGSEE148 pKa = 3.94TVTTIVKK155 pKa = 10.13DD156 pKa = 3.46PSGTYY161 pKa = 9.59TYY163 pKa = 10.57TNEE166 pKa = 4.03AGVAMVIDD174 pKa = 3.89VQADD178 pKa = 3.49VVNNFEE184 pKa = 4.36EE185 pKa = 4.91IINKK189 pKa = 8.56TEE191 pKa = 3.94VQEE194 pKa = 5.68LLTQVINNNGGNVHH208 pKa = 6.46FDD210 pKa = 3.22EE211 pKa = 6.44GGFTYY216 pKa = 10.37TDD218 pKa = 3.47EE219 pKa = 4.69NGASQTVDD227 pKa = 2.82LTTIVQGSEE236 pKa = 3.94TVTTIVKK243 pKa = 10.13DD244 pKa = 3.46PSGTYY249 pKa = 9.59TYY251 pKa = 10.57TNEE254 pKa = 4.03AGVAMVIDD262 pKa = 3.89VQADD266 pKa = 3.49VVNNFEE272 pKa = 4.36EE273 pKa = 4.91IINKK277 pKa = 8.56TEE279 pKa = 3.94VQEE282 pKa = 5.68LLTQVINNNGGNVHH296 pKa = 6.46FDD298 pKa = 3.22EE299 pKa = 6.44GGFTYY304 pKa = 10.37TDD306 pKa = 3.47EE307 pKa = 4.69NGASQTVDD315 pKa = 2.82LTTIVQGSEE324 pKa = 3.94TVTTIVKK331 pKa = 10.13DD332 pKa = 3.46PSGTYY337 pKa = 9.59TYY339 pKa = 10.57TNEE342 pKa = 4.03AGVAMVIDD350 pKa = 3.89VQADD354 pKa = 3.49VVNNFEE360 pKa = 4.36EE361 pKa = 4.91IINKK365 pKa = 8.56TEE367 pKa = 3.94VQEE370 pKa = 5.68LLTQVINNNGGNVHH384 pKa = 6.46FDD386 pKa = 3.22EE387 pKa = 6.44GGFTYY392 pKa = 10.37TDD394 pKa = 3.47EE395 pKa = 4.69NGASQTVDD403 pKa = 2.82LTTIVQGSEE412 pKa = 3.94TVTTIVKK419 pKa = 10.13DD420 pKa = 3.46PSGTYY425 pKa = 9.59TYY427 pKa = 10.57TNEE430 pKa = 3.8AGVAVVIDD438 pKa = 3.68VQADD442 pKa = 3.52VVNNFEE448 pKa = 4.36EE449 pKa = 4.91IINKK453 pKa = 8.56TEE455 pKa = 3.94VQEE458 pKa = 5.68LLTQVINNNGGNVHH472 pKa = 6.46FDD474 pKa = 3.22EE475 pKa = 6.44GGFTYY480 pKa = 10.37TDD482 pKa = 3.47EE483 pKa = 4.69NGASQTVDD491 pKa = 2.82LTTIVQGSEE500 pKa = 3.94TVTTIVKK507 pKa = 10.13DD508 pKa = 3.46PSGTYY513 pKa = 9.59TYY515 pKa = 10.57TNEE518 pKa = 4.03AGVAMVIDD526 pKa = 3.89VQADD530 pKa = 3.49VVNNFEE536 pKa = 4.36EE537 pKa = 4.91IINKK541 pKa = 8.56TEE543 pKa = 3.94VQEE546 pKa = 5.36LLTQVVNNVGGNMTYY561 pKa = 10.83DD562 pKa = 3.47GTGFTYY568 pKa = 10.63TDD570 pKa = 3.61GNGDD574 pKa = 3.81TVAMDD579 pKa = 3.48MSTFVQGNEE588 pKa = 3.87TTTTLVKK595 pKa = 10.57DD596 pKa = 3.57ASDD599 pKa = 3.52NGKK602 pKa = 8.5YY603 pKa = 9.22TYY605 pKa = 10.52TNEE608 pKa = 4.02TNTTVEE614 pKa = 4.15LDD616 pKa = 3.23IPTDD620 pKa = 3.53VINNFEE626 pKa = 4.63TIVADD631 pKa = 3.95ANVGNLLQQLVVKK644 pKa = 10.73SNTLVEE650 pKa = 4.15TAVNYY655 pKa = 10.06VVLDD659 pKa = 3.45VDD661 pKa = 3.49TTIIVDD667 pKa = 3.87AEE669 pKa = 4.16AADD672 pKa = 3.85VTITLPAATSEE683 pKa = 4.17NKK685 pKa = 10.17GRR687 pKa = 11.84MITIRR692 pKa = 11.84KK693 pKa = 8.81IDD695 pKa = 3.63EE696 pKa = 3.97SDD698 pKa = 3.26NAVNFSQQIKK708 pKa = 7.96TSKK711 pKa = 9.75TEE713 pKa = 3.84GFTSINFGITVNIQSNGSVWYY734 pKa = 8.63MINN737 pKa = 3.18
MM1 pKa = 7.49TNNFEE6 pKa = 4.36EE7 pKa = 4.86IINRR11 pKa = 11.84TEE13 pKa = 3.82VQEE16 pKa = 4.11VLNQYY21 pKa = 8.08ITEE24 pKa = 4.22NGGNIHH30 pKa = 6.63YY31 pKa = 10.48DD32 pKa = 3.3GGSFTYY38 pKa = 10.63LDD40 pKa = 3.86EE41 pKa = 4.87NGNTVTMDD49 pKa = 3.3MTTIIQGSEE58 pKa = 4.0TVTTLVKK65 pKa = 10.84DD66 pKa = 3.51PLNNGKK72 pKa = 10.23YY73 pKa = 9.93IYY75 pKa = 9.73TNEE78 pKa = 4.07AGVEE82 pKa = 4.18VVIDD86 pKa = 3.51VTTEE90 pKa = 3.51VTNNFEE96 pKa = 4.27EE97 pKa = 4.81IINRR101 pKa = 11.84TEE103 pKa = 3.82VQEE106 pKa = 4.11VLNQYY111 pKa = 8.08ITEE114 pKa = 4.22NGGNIHH120 pKa = 6.63YY121 pKa = 10.48DD122 pKa = 3.3GGSFTYY128 pKa = 10.63LDD130 pKa = 3.86EE131 pKa = 4.87NGNTVTMDD139 pKa = 3.02MTTIVQGSEE148 pKa = 3.94TVTTIVKK155 pKa = 10.13DD156 pKa = 3.46PSGTYY161 pKa = 9.59TYY163 pKa = 10.57TNEE166 pKa = 4.03AGVAMVIDD174 pKa = 3.89VQADD178 pKa = 3.49VVNNFEE184 pKa = 4.36EE185 pKa = 4.91IINKK189 pKa = 8.56TEE191 pKa = 3.94VQEE194 pKa = 5.68LLTQVINNNGGNVHH208 pKa = 6.46FDD210 pKa = 3.22EE211 pKa = 6.44GGFTYY216 pKa = 10.37TDD218 pKa = 3.47EE219 pKa = 4.69NGASQTVDD227 pKa = 2.82LTTIVQGSEE236 pKa = 3.94TVTTIVKK243 pKa = 10.13DD244 pKa = 3.46PSGTYY249 pKa = 9.59TYY251 pKa = 10.57TNEE254 pKa = 4.03AGVAMVIDD262 pKa = 3.89VQADD266 pKa = 3.49VVNNFEE272 pKa = 4.36EE273 pKa = 4.91IINKK277 pKa = 8.56TEE279 pKa = 3.94VQEE282 pKa = 5.68LLTQVINNNGGNVHH296 pKa = 6.46FDD298 pKa = 3.22EE299 pKa = 6.44GGFTYY304 pKa = 10.37TDD306 pKa = 3.47EE307 pKa = 4.69NGASQTVDD315 pKa = 2.82LTTIVQGSEE324 pKa = 3.94TVTTIVKK331 pKa = 10.13DD332 pKa = 3.46PSGTYY337 pKa = 9.59TYY339 pKa = 10.57TNEE342 pKa = 4.03AGVAMVIDD350 pKa = 3.89VQADD354 pKa = 3.49VVNNFEE360 pKa = 4.36EE361 pKa = 4.91IINKK365 pKa = 8.56TEE367 pKa = 3.94VQEE370 pKa = 5.68LLTQVINNNGGNVHH384 pKa = 6.46FDD386 pKa = 3.22EE387 pKa = 6.44GGFTYY392 pKa = 10.37TDD394 pKa = 3.47EE395 pKa = 4.69NGASQTVDD403 pKa = 2.82LTTIVQGSEE412 pKa = 3.94TVTTIVKK419 pKa = 10.13DD420 pKa = 3.46PSGTYY425 pKa = 9.59TYY427 pKa = 10.57TNEE430 pKa = 3.8AGVAVVIDD438 pKa = 3.68VQADD442 pKa = 3.52VVNNFEE448 pKa = 4.36EE449 pKa = 4.91IINKK453 pKa = 8.56TEE455 pKa = 3.94VQEE458 pKa = 5.68LLTQVINNNGGNVHH472 pKa = 6.46FDD474 pKa = 3.22EE475 pKa = 6.44GGFTYY480 pKa = 10.37TDD482 pKa = 3.47EE483 pKa = 4.69NGASQTVDD491 pKa = 2.82LTTIVQGSEE500 pKa = 3.94TVTTIVKK507 pKa = 10.13DD508 pKa = 3.46PSGTYY513 pKa = 9.59TYY515 pKa = 10.57TNEE518 pKa = 4.03AGVAMVIDD526 pKa = 3.89VQADD530 pKa = 3.49VVNNFEE536 pKa = 4.36EE537 pKa = 4.91IINKK541 pKa = 8.56TEE543 pKa = 3.94VQEE546 pKa = 5.36LLTQVVNNVGGNMTYY561 pKa = 10.83DD562 pKa = 3.47GTGFTYY568 pKa = 10.63TDD570 pKa = 3.61GNGDD574 pKa = 3.81TVAMDD579 pKa = 3.48MSTFVQGNEE588 pKa = 3.87TTTTLVKK595 pKa = 10.57DD596 pKa = 3.57ASDD599 pKa = 3.52NGKK602 pKa = 8.5YY603 pKa = 9.22TYY605 pKa = 10.52TNEE608 pKa = 4.02TNTTVEE614 pKa = 4.15LDD616 pKa = 3.23IPTDD620 pKa = 3.53VINNFEE626 pKa = 4.63TIVADD631 pKa = 3.95ANVGNLLQQLVVKK644 pKa = 10.73SNTLVEE650 pKa = 4.15TAVNYY655 pKa = 10.06VVLDD659 pKa = 3.45VDD661 pKa = 3.49TTIIVDD667 pKa = 3.87AEE669 pKa = 4.16AADD672 pKa = 3.85VTITLPAATSEE683 pKa = 4.17NKK685 pKa = 10.17GRR687 pKa = 11.84MITIRR692 pKa = 11.84KK693 pKa = 8.81IDD695 pKa = 3.63EE696 pKa = 3.97SDD698 pKa = 3.26NAVNFSQQIKK708 pKa = 7.96TSKK711 pKa = 9.75TEE713 pKa = 3.84GFTSINFGITVNIQSNGSVWYY734 pKa = 8.63MINN737 pKa = 3.18
Molecular weight: 80.03 kDa
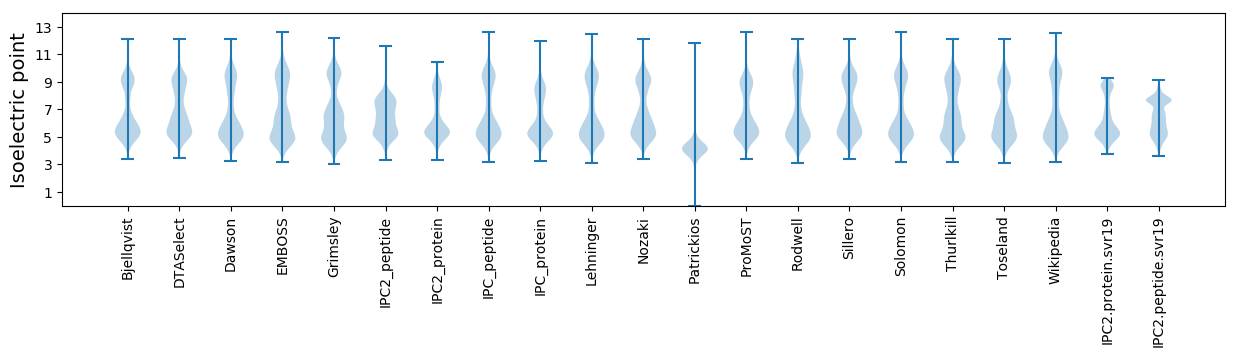
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H1Z845|H1Z845_MYROD Quinolinate phosphoribosyltransferase [decarboxylating] OS=Myroides odoratus DSM 2801 OX=929704 GN=Myrod_0998 PE=3 SV=1
MM1 pKa = 7.31NRR3 pKa = 11.84FSLSFLRR10 pKa = 11.84SIAFLFLVVGMILMVQNIDD29 pKa = 3.23QFLYY33 pKa = 10.53KK34 pKa = 9.1QTLEE38 pKa = 3.92IGSLAFALACITISILGQRR57 pKa = 11.84KK58 pKa = 8.7LKK60 pKa = 10.43KK61 pKa = 8.82STRR64 pKa = 11.84ISNLL68 pKa = 3.06
MM1 pKa = 7.31NRR3 pKa = 11.84FSLSFLRR10 pKa = 11.84SIAFLFLVVGMILMVQNIDD29 pKa = 3.23QFLYY33 pKa = 10.53KK34 pKa = 9.1QTLEE38 pKa = 3.92IGSLAFALACITISILGQRR57 pKa = 11.84KK58 pKa = 8.7LKK60 pKa = 10.43KK61 pKa = 8.82STRR64 pKa = 11.84ISNLL68 pKa = 3.06
Molecular weight: 7.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1212748 |
30 |
2726 |
328.9 |
37.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.721 ± 0.043 | 0.761 ± 0.011 |
5.268 ± 0.03 | 6.653 ± 0.044 |
5.061 ± 0.03 | 6.284 ± 0.042 |
1.867 ± 0.019 | 7.546 ± 0.04 |
7.107 ± 0.042 | 9.558 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.257 ± 0.02 | 5.549 ± 0.046 |
3.324 ± 0.023 | 4.283 ± 0.031 |
3.477 ± 0.027 | 5.952 ± 0.029 |
6.158 ± 0.051 | 6.68 ± 0.036 |
1.042 ± 0.013 | 4.454 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
