
Northern cereal mosaic virus (NCMV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Cytorhabdovirus
Average proteome isoelectric point is 7.02
Get precalculated fractions of proteins
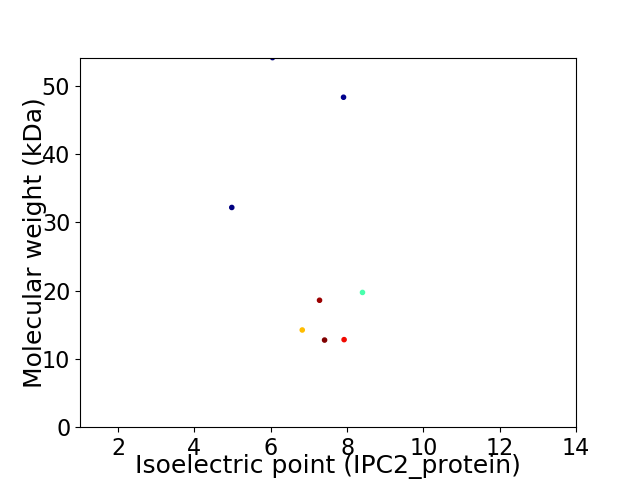
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q9JGU1|NCAP_NCMV Nucleoprotein OS=Northern cereal mosaic virus OX=1985704 GN=N PE=3 SV=1
MM1 pKa = 7.94DD2 pKa = 4.9KK3 pKa = 10.38KK4 pKa = 10.99ASGISGEE11 pKa = 4.06NALFGDD17 pKa = 3.99VPNDD21 pKa = 3.63VVGTTYY27 pKa = 11.37EE28 pKa = 4.05MGLDD32 pKa = 4.28GIFDD36 pKa = 4.37DD37 pKa = 5.89GEE39 pKa = 4.26TDD41 pKa = 4.03VIEE44 pKa = 4.73SPADD48 pKa = 3.48AEE50 pKa = 4.37EE51 pKa = 4.52HH52 pKa = 4.93VTTDD56 pKa = 2.77IVADD60 pKa = 3.98EE61 pKa = 4.59GDD63 pKa = 3.65NLITKK68 pKa = 9.45VDD70 pKa = 3.72DD71 pKa = 3.84MIGYY75 pKa = 9.26LKK77 pKa = 10.51RR78 pKa = 11.84EE79 pKa = 4.29CQDD82 pKa = 2.58HH83 pKa = 6.5GIAVRR88 pKa = 11.84KK89 pKa = 9.12EE90 pKa = 3.8WVNVLTRR97 pKa = 11.84KK98 pKa = 9.27FHH100 pKa = 5.47MSKK103 pKa = 10.45KK104 pKa = 9.83IYY106 pKa = 10.2ASHH109 pKa = 7.52LDD111 pKa = 3.7FFLLGVMGEE120 pKa = 3.88RR121 pKa = 11.84HH122 pKa = 5.04VAVEE126 pKa = 4.53KK127 pKa = 10.77DD128 pKa = 3.85FKK130 pKa = 10.57DD131 pKa = 3.37TAVRR135 pKa = 11.84LSDD138 pKa = 4.01EE139 pKa = 4.34VNKK142 pKa = 10.32MSGISKK148 pKa = 10.05KK149 pKa = 10.63VADD152 pKa = 4.24NEE154 pKa = 4.0TRR156 pKa = 11.84MIKK159 pKa = 10.39EE160 pKa = 4.32LDD162 pKa = 3.38AKK164 pKa = 8.94MKK166 pKa = 10.68EE167 pKa = 3.95MGSLIGKK174 pKa = 9.26FNGVLNEE181 pKa = 4.41FKK183 pKa = 11.03GSMAVSSRR191 pKa = 11.84PASVASWALDD201 pKa = 3.21QTTEE205 pKa = 3.69TSRR208 pKa = 11.84EE209 pKa = 3.68KK210 pKa = 10.94NYY212 pKa = 11.11NEE214 pKa = 3.82FLKK217 pKa = 11.08KK218 pKa = 10.56LGFQDD223 pKa = 3.6HH224 pKa = 6.91HH225 pKa = 7.04IKK227 pKa = 10.53SPLMKK232 pKa = 9.7KK233 pKa = 8.11CTVMITDD240 pKa = 3.52EE241 pKa = 4.43MYY243 pKa = 11.23DD244 pKa = 3.42EE245 pKa = 4.93VMLEE249 pKa = 4.05NTSEE253 pKa = 4.06DD254 pKa = 3.25TLGIYY259 pKa = 9.97KK260 pKa = 9.71EE261 pKa = 4.01QIITYY266 pKa = 8.09GQSIQKK272 pKa = 10.18KK273 pKa = 6.86VEE275 pKa = 3.84KK276 pKa = 9.74PIKK279 pKa = 9.33KK280 pKa = 10.16DD281 pKa = 3.25PYY283 pKa = 11.25ADD285 pKa = 3.64FF286 pKa = 5.14
MM1 pKa = 7.94DD2 pKa = 4.9KK3 pKa = 10.38KK4 pKa = 10.99ASGISGEE11 pKa = 4.06NALFGDD17 pKa = 3.99VPNDD21 pKa = 3.63VVGTTYY27 pKa = 11.37EE28 pKa = 4.05MGLDD32 pKa = 4.28GIFDD36 pKa = 4.37DD37 pKa = 5.89GEE39 pKa = 4.26TDD41 pKa = 4.03VIEE44 pKa = 4.73SPADD48 pKa = 3.48AEE50 pKa = 4.37EE51 pKa = 4.52HH52 pKa = 4.93VTTDD56 pKa = 2.77IVADD60 pKa = 3.98EE61 pKa = 4.59GDD63 pKa = 3.65NLITKK68 pKa = 9.45VDD70 pKa = 3.72DD71 pKa = 3.84MIGYY75 pKa = 9.26LKK77 pKa = 10.51RR78 pKa = 11.84EE79 pKa = 4.29CQDD82 pKa = 2.58HH83 pKa = 6.5GIAVRR88 pKa = 11.84KK89 pKa = 9.12EE90 pKa = 3.8WVNVLTRR97 pKa = 11.84KK98 pKa = 9.27FHH100 pKa = 5.47MSKK103 pKa = 10.45KK104 pKa = 9.83IYY106 pKa = 10.2ASHH109 pKa = 7.52LDD111 pKa = 3.7FFLLGVMGEE120 pKa = 3.88RR121 pKa = 11.84HH122 pKa = 5.04VAVEE126 pKa = 4.53KK127 pKa = 10.77DD128 pKa = 3.85FKK130 pKa = 10.57DD131 pKa = 3.37TAVRR135 pKa = 11.84LSDD138 pKa = 4.01EE139 pKa = 4.34VNKK142 pKa = 10.32MSGISKK148 pKa = 10.05KK149 pKa = 10.63VADD152 pKa = 4.24NEE154 pKa = 4.0TRR156 pKa = 11.84MIKK159 pKa = 10.39EE160 pKa = 4.32LDD162 pKa = 3.38AKK164 pKa = 8.94MKK166 pKa = 10.68EE167 pKa = 3.95MGSLIGKK174 pKa = 9.26FNGVLNEE181 pKa = 4.41FKK183 pKa = 11.03GSMAVSSRR191 pKa = 11.84PASVASWALDD201 pKa = 3.21QTTEE205 pKa = 3.69TSRR208 pKa = 11.84EE209 pKa = 3.68KK210 pKa = 10.94NYY212 pKa = 11.11NEE214 pKa = 3.82FLKK217 pKa = 11.08KK218 pKa = 10.56LGFQDD223 pKa = 3.6HH224 pKa = 6.91HH225 pKa = 7.04IKK227 pKa = 10.53SPLMKK232 pKa = 9.7KK233 pKa = 8.11CTVMITDD240 pKa = 3.52EE241 pKa = 4.43MYY243 pKa = 11.23DD244 pKa = 3.42EE245 pKa = 4.93VMLEE249 pKa = 4.05NTSEE253 pKa = 4.06DD254 pKa = 3.25TLGIYY259 pKa = 9.97KK260 pKa = 9.71EE261 pKa = 4.01QIITYY266 pKa = 8.09GQSIQKK272 pKa = 10.18KK273 pKa = 6.86VEE275 pKa = 3.84KK276 pKa = 9.74PIKK279 pKa = 9.33KK280 pKa = 10.16DD281 pKa = 3.25PYY283 pKa = 11.25ADD285 pKa = 3.64FF286 pKa = 5.14
Molecular weight: 32.17 kDa
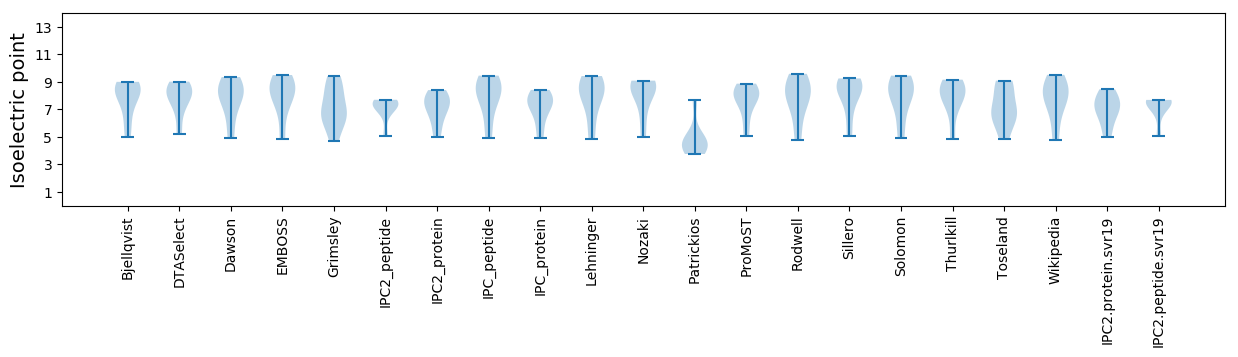
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9JGT6|VP6_NCMV Protein 6 OS=Northern cereal mosaic virus OX=1985704 GN=6 PE=4 SV=1
MM1 pKa = 6.88ATEE4 pKa = 3.98VSTRR8 pKa = 11.84FIFIKK13 pKa = 10.04IDD15 pKa = 3.45ALSTLHH21 pKa = 7.17FIDD24 pKa = 3.83EE25 pKa = 4.58GGALSMNGDD34 pKa = 3.95SIEE37 pKa = 3.9FLIGKK42 pKa = 9.77SSLDD46 pKa = 3.13KK47 pKa = 11.29GLLKK51 pKa = 10.56RR52 pKa = 11.84ATKK55 pKa = 10.61LFTWLFSPEE64 pKa = 3.5IAKK67 pKa = 9.39QHH69 pKa = 5.81LSITRR74 pKa = 11.84STEE77 pKa = 3.83SNVYY81 pKa = 7.85MTDD84 pKa = 2.82ATRR87 pKa = 11.84YY88 pKa = 9.19KK89 pKa = 10.8YY90 pKa = 10.28IFPEE94 pKa = 4.09YY95 pKa = 10.73LLVRR99 pKa = 11.84YY100 pKa = 9.75VGTKK104 pKa = 9.88FPDD107 pKa = 3.28MKK109 pKa = 10.34IVADD113 pKa = 3.57ATFKK117 pKa = 10.77RR118 pKa = 11.84RR119 pKa = 11.84NPKK122 pKa = 10.22GDD124 pKa = 3.75VIGMLDD130 pKa = 3.64LSFKK134 pKa = 8.96EE135 pKa = 4.09VRR137 pKa = 11.84VKK139 pKa = 10.47EE140 pKa = 4.14VSEE143 pKa = 4.28TKK145 pKa = 10.42AIEE148 pKa = 4.19LRR150 pKa = 11.84DD151 pKa = 3.99SNPGYY156 pKa = 10.7LLSTTYY162 pKa = 10.44RR163 pKa = 11.84EE164 pKa = 4.43SEE166 pKa = 4.28SLPVVSKK173 pKa = 10.52PP174 pKa = 3.48
MM1 pKa = 6.88ATEE4 pKa = 3.98VSTRR8 pKa = 11.84FIFIKK13 pKa = 10.04IDD15 pKa = 3.45ALSTLHH21 pKa = 7.17FIDD24 pKa = 3.83EE25 pKa = 4.58GGALSMNGDD34 pKa = 3.95SIEE37 pKa = 3.9FLIGKK42 pKa = 9.77SSLDD46 pKa = 3.13KK47 pKa = 11.29GLLKK51 pKa = 10.56RR52 pKa = 11.84ATKK55 pKa = 10.61LFTWLFSPEE64 pKa = 3.5IAKK67 pKa = 9.39QHH69 pKa = 5.81LSITRR74 pKa = 11.84STEE77 pKa = 3.83SNVYY81 pKa = 7.85MTDD84 pKa = 2.82ATRR87 pKa = 11.84YY88 pKa = 9.19KK89 pKa = 10.8YY90 pKa = 10.28IFPEE94 pKa = 4.09YY95 pKa = 10.73LLVRR99 pKa = 11.84YY100 pKa = 9.75VGTKK104 pKa = 9.88FPDD107 pKa = 3.28MKK109 pKa = 10.34IVADD113 pKa = 3.57ATFKK117 pKa = 10.77RR118 pKa = 11.84RR119 pKa = 11.84NPKK122 pKa = 10.22GDD124 pKa = 3.75VIGMLDD130 pKa = 3.64LSFKK134 pKa = 8.96EE135 pKa = 4.09VRR137 pKa = 11.84VKK139 pKa = 10.47EE140 pKa = 4.14VSEE143 pKa = 4.28TKK145 pKa = 10.42AIEE148 pKa = 4.19LRR150 pKa = 11.84DD151 pKa = 3.99SNPGYY156 pKa = 10.7LLSTTYY162 pKa = 10.44RR163 pKa = 11.84EE164 pKa = 4.43SEE166 pKa = 4.28SLPVVSKK173 pKa = 10.52PP174 pKa = 3.48
Molecular weight: 19.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1907 |
114 |
483 |
238.4 |
26.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.611 ± 0.728 | 1.888 ± 0.611 |
5.716 ± 0.817 | 6.502 ± 0.448 |
4.038 ± 0.244 | 6.45 ± 0.372 |
1.468 ± 0.369 | 7.184 ± 0.732 |
7.446 ± 0.664 | 8.233 ± 0.678 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.937 ± 0.498 | 3.671 ± 0.373 |
3.776 ± 0.506 | 2.832 ± 0.592 |
4.615 ± 0.622 | 9.229 ± 0.809 |
6.24 ± 0.275 | 7.918 ± 0.72 |
1.206 ± 0.27 | 3.041 ± 0.448 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
