
Sphingobacterium wenxiniae
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Sphingobacteriia; Sphingobacteriales; Sphingobacteriaceae; Sphingobacterium
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
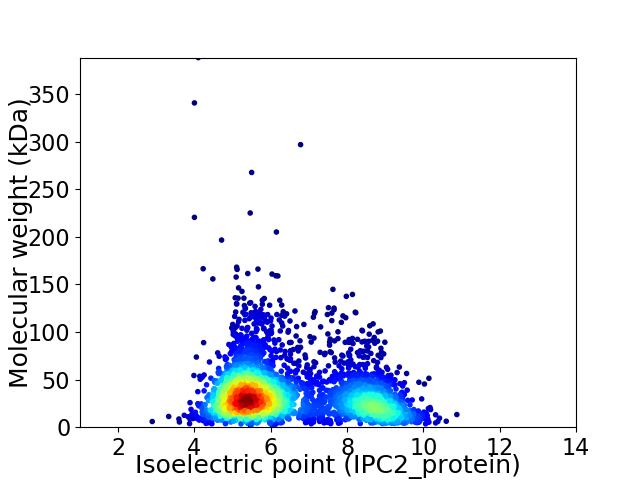
Virtual 2D-PAGE plot for 3412 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I6Q3Y3|A0A1I6Q3Y3_9SPHI Starch-binding associating with outer membrane OS=Sphingobacterium wenxiniae OX=683125 GN=SAMN05660206_102103 PE=3 SV=1
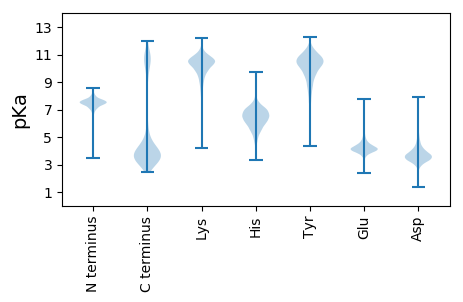
MM1 pKa = 7.48KK2 pKa = 10.11KK3 pKa = 9.56IRR5 pKa = 11.84YY6 pKa = 8.81ILISLHH12 pKa = 6.8LVLLLLGGISSLHH25 pKa = 5.58AQCVDD30 pKa = 3.24NTGLIAFTGYY40 pKa = 10.55QIADD44 pKa = 3.28AGNPQGQSDD53 pKa = 3.92RR54 pKa = 11.84FSFVTLDD61 pKa = 3.9SIPPNTPIRR70 pKa = 11.84FTDD73 pKa = 3.09RR74 pKa = 11.84GFTGGSSFQDD84 pKa = 3.31FGNALTDD91 pKa = 4.65GDD93 pKa = 4.85FSWSDD98 pKa = 3.27ASGISPGTHH107 pKa = 4.49IVVTLGSGTATSTIGSITGYY127 pKa = 10.4LGNFNLGLSGDD138 pKa = 3.77QIFAYY143 pKa = 8.62TASPSFKK150 pKa = 9.61IHH152 pKa = 6.73AAMLINKK159 pKa = 7.29SAWDD163 pKa = 3.76TEE165 pKa = 4.05MLEE168 pKa = 4.23LGNPIVTSSKK178 pKa = 10.78SLNPAIAPVRR188 pKa = 11.84QSITVYY194 pKa = 10.96NSTDD198 pKa = 3.23DD199 pKa = 4.07AFQAKK204 pKa = 9.85LNPITLTGSRR214 pKa = 11.84TALRR218 pKa = 11.84NQLFAGIFNIANSLNTDD235 pKa = 3.19AVNLSVSPLTWNASVPTYY253 pKa = 10.74TPISLNNKK261 pKa = 8.59ILSSTVTDD269 pKa = 2.99ACYY272 pKa = 10.24IQWQSSIDD280 pKa = 3.88GINFSNINDD289 pKa = 3.56GTAFSGTKK297 pKa = 8.68TASLTINTPLVADD310 pKa = 3.77TWFRR314 pKa = 11.84MHH316 pKa = 6.74LTGAAYY322 pKa = 8.48TVTDD326 pKa = 5.33AIKK329 pKa = 9.35LTLPAVTAHH338 pKa = 6.77PLNRR342 pKa = 11.84SICVGEE348 pKa = 3.94NTSFSVTATGTDD360 pKa = 3.66LQYY363 pKa = 11.02QWQEE367 pKa = 3.73DD368 pKa = 3.85RR369 pKa = 11.84GSGFTDD375 pKa = 3.89LSDD378 pKa = 3.35GGVYY382 pKa = 10.41SGVTTSTLNISGATAGMDD400 pKa = 3.95GYY402 pKa = 10.37QYY404 pKa = 10.81RR405 pKa = 11.84VRR407 pKa = 11.84VYY409 pKa = 10.91SSIANPVTSNSATLSVSNIQATISKK434 pKa = 9.15TDD436 pKa = 3.21VGCFGEE442 pKa = 4.44ASGSATVSNVMGGIGSYY459 pKa = 8.9TYY461 pKa = 10.33SWVRR465 pKa = 11.84QGTTDD470 pKa = 3.92IISDD474 pKa = 3.85TEE476 pKa = 4.23TASGLTEE483 pKa = 3.7GTYY486 pKa = 10.1TVTIRR491 pKa = 11.84DD492 pKa = 3.72AIGCVKK498 pKa = 9.68TLDD501 pKa = 4.03VIIMQPSALTASTTQTNVSCFNGSNGTATVSVSGGTGPYY540 pKa = 8.7TYY542 pKa = 10.73SWLPSGGTSQTATGLSAGTYY562 pKa = 7.78TVTVTDD568 pKa = 4.67ANSCQITRR576 pKa = 11.84SFTITHH582 pKa = 7.15PPALMASDD590 pKa = 3.96GGKK593 pKa = 8.73TNVSCNGGSNGTATVNVVGGTGAYY617 pKa = 8.11TYY619 pKa = 9.18TYY621 pKa = 9.87SWAPFGGTSQTATGLSAGLYY641 pKa = 8.09TVTVTDD647 pKa = 4.59ANGCQTTQNFEE658 pKa = 3.77ISEE661 pKa = 4.22PTSAVVITTSDD672 pKa = 2.93IMHH675 pKa = 6.66VSTVGGSDD683 pKa = 3.21GSATVDD689 pKa = 3.36VMGGTAPYY697 pKa = 8.8TYY699 pKa = 10.06SWSPTGGTGATATGLSAGVYY719 pKa = 8.4TVTVTDD725 pKa = 5.15ANTCTYY731 pKa = 9.51SHH733 pKa = 6.68NVTIEE738 pKa = 3.91QQAVNQAPVISNLNGDD754 pKa = 3.63AVTFTEE760 pKa = 5.2DD761 pKa = 3.25GPAVLLDD768 pKa = 3.45VFGNATVSDD777 pKa = 4.37PDD779 pKa = 3.74SPDD782 pKa = 3.38FNEE785 pKa = 4.23GNLTVSIVTNGVAAEE800 pKa = 4.0DD801 pKa = 3.6RR802 pKa = 11.84LGIRR806 pKa = 11.84HH807 pKa = 6.3EE808 pKa = 4.5GSGPGLIDD816 pKa = 3.31VAGGDD821 pKa = 3.8VYY823 pKa = 11.64YY824 pKa = 10.75GGTPIGSLTGGIAGQDD840 pKa = 3.25LVVTFLPNSTPAAAQALIRR859 pKa = 11.84NLTYY863 pKa = 11.39SNVNTGNPSSAPRR876 pKa = 11.84TVYY879 pKa = 8.89ITLSDD884 pKa = 3.78GDD886 pKa = 4.25GGTSAVQAVTVSIKK900 pKa = 9.55TVNDD904 pKa = 3.28APTISVPVQINVTEE918 pKa = 4.25DD919 pKa = 3.35VPVALTGITFYY930 pKa = 11.22DD931 pKa = 3.45VDD933 pKa = 3.92NTDD936 pKa = 2.92VTVTFRR942 pKa = 11.84TTIAGAGSGTLTATSTADD960 pKa = 3.33VTVSGSGTNEE970 pKa = 3.43LTMAGPWASINNLIAANGLVFLTDD994 pKa = 3.32QDD996 pKa = 3.8NVNTVFLNIQVSDD1009 pKa = 3.83GALADD1014 pKa = 3.73STATSLIVAAVNDD1027 pKa = 4.09APQISVPLLQTVLQDD1042 pKa = 3.25ATLTFNAANGNPISISDD1059 pKa = 3.58VDD1061 pKa = 4.34AGTNPIKK1068 pKa = 11.01LNLTTTRR1075 pKa = 11.84GLLSLGSTNGLSFDD1089 pKa = 4.28IGNGTADD1096 pKa = 3.18NTMEE1100 pKa = 4.25FTGTLADD1107 pKa = 4.31INAALNGLAFEE1118 pKa = 4.55PTGGYY1123 pKa = 9.51YY1124 pKa = 10.57GPAAINITADD1134 pKa = 3.51DD1135 pKa = 3.77QGATGAGGPKK1145 pKa = 9.87SVTRR1149 pKa = 11.84EE1150 pKa = 3.47IPITVEE1156 pKa = 3.45PLNPKK1161 pKa = 7.98VTSVSASTADD1171 pKa = 3.65GLYY1174 pKa = 10.64KK1175 pKa = 10.38ADD1177 pKa = 4.61DD1178 pKa = 4.59VITLTVTFNQNVVVNTNGGDD1198 pKa = 3.57TPTLLLEE1205 pKa = 4.11TGIIDD1210 pKa = 4.16RR1211 pKa = 11.84SAIYY1215 pKa = 9.77MGGSGSNTLTFIYY1228 pKa = 9.41TVQPGDD1234 pKa = 3.3VSADD1238 pKa = 3.35LDD1240 pKa = 4.13YY1241 pKa = 11.8VSISALSLNGATILSSYY1258 pKa = 10.51NGANAVLTLPTVGGASSIAGQKK1280 pKa = 10.51NIVIDD1285 pKa = 4.54GIVPVVNAVAVPANAYY1301 pKa = 9.63YY1302 pKa = 10.94GSGDD1306 pKa = 3.3QLYY1309 pKa = 7.98FTVEE1313 pKa = 3.92FSEE1316 pKa = 4.38AVAVNTTGGSPALSLTIGSQTVSAQYY1342 pKa = 9.05LTEE1345 pKa = 4.36SSTTTDD1351 pKa = 3.19LLFRR1355 pKa = 11.84YY1356 pKa = 9.27SVAAGMQDD1364 pKa = 2.72HH1365 pKa = 7.46DD1366 pKa = 4.9GIEE1369 pKa = 4.07IGTLSLNGATIRR1381 pKa = 11.84DD1382 pKa = 3.65AAGNDD1387 pKa = 3.48AVLTLNNVGATTHH1400 pKa = 5.63VKK1402 pKa = 10.04VDD1404 pKa = 3.36AKK1406 pKa = 11.08SPVVTMTINGGATHH1420 pKa = 6.53TNDD1423 pKa = 3.65RR1424 pKa = 11.84NVTLTLTTADD1434 pKa = 3.33TDD1436 pKa = 3.87VATMSFMNSPIFFPGSGASQPEE1458 pKa = 4.03PFAPTKK1464 pKa = 10.3AWQLTAGDD1472 pKa = 3.69GMKK1475 pKa = 9.97FVSGIVIDD1483 pKa = 4.04GAGNLTSVLAVITLDD1498 pKa = 3.1QTAPIVTGVDD1508 pKa = 2.82EE1509 pKa = 4.88GEE1511 pKa = 4.48SYY1513 pKa = 9.37NTDD1516 pKa = 2.8RR1517 pKa = 11.84TITFNEE1523 pKa = 4.26GTATLNGATFISGTTVSQDD1542 pKa = 2.77GAYY1545 pKa = 9.12TLIVTDD1551 pKa = 3.64EE1552 pKa = 4.75AGNSTTLSFAIDD1564 pKa = 3.35QTAPAVPAGLTATPDD1579 pKa = 3.14AGNINLQWTANQEE1592 pKa = 3.93ADD1594 pKa = 3.18LAAYY1598 pKa = 10.09RR1599 pKa = 11.84LYY1601 pKa = 11.0SGTAADD1607 pKa = 4.0QLSVLADD1614 pKa = 3.65IPAGTTTYY1622 pKa = 9.45THH1624 pKa = 6.96GPLTMGQRR1632 pKa = 11.84YY1633 pKa = 9.05YY1634 pKa = 11.24YY1635 pKa = 10.52AITAIDD1641 pKa = 3.5ALGNEE1646 pKa = 4.75SAQSVVVDD1654 pKa = 5.08ALPQDD1659 pKa = 3.76TQTITFGTLNDD1670 pKa = 3.75MTYY1673 pKa = 11.38GDD1675 pKa = 4.77APFALTATTTSGLTVAYY1692 pKa = 9.92SSSDD1696 pKa = 3.26NSIASIAGDD1705 pKa = 3.51VLTIHH1710 pKa = 6.18QAGDD1714 pKa = 3.54VVITASQNGNNAFLAATPVTQALHH1738 pKa = 5.9INKK1741 pKa = 9.57ADD1743 pKa = 3.68FAGITFDD1750 pKa = 5.28DD1751 pKa = 3.86KK1752 pKa = 11.26TVTYY1756 pKa = 10.19DD1757 pKa = 3.76GSAHH1761 pKa = 6.8SIFVSNAPTDD1771 pKa = 3.39ATVIYY1776 pKa = 9.97TNNNQINAGTYY1787 pKa = 7.96TVMATVSQANYY1798 pKa = 10.5NDD1800 pKa = 4.14LPLTAEE1806 pKa = 4.12LTIEE1810 pKa = 4.16KK1811 pKa = 10.35ADD1813 pKa = 3.72FAGITFNDD1821 pKa = 3.25ATVTYY1826 pKa = 9.67DD1827 pKa = 3.8GSAHH1831 pKa = 6.51SIAVANTPTGATITYY1846 pKa = 9.54VGNAQINAGTYY1857 pKa = 6.69TVRR1860 pKa = 11.84ATVSQANYY1868 pKa = 10.1NDD1870 pKa = 3.87LVLTAEE1876 pKa = 4.31LEE1878 pKa = 4.07ITKK1881 pKa = 10.97AMLTDD1886 pKa = 3.23IALRR1890 pKa = 11.84NNSFVYY1896 pKa = 10.57DD1897 pKa = 4.04GNAKK1901 pKa = 10.08SLEE1904 pKa = 3.95ISGILPTGVQVIYY1917 pKa = 10.17TGNNQVNAGMYY1928 pKa = 8.74TVVAEE1933 pKa = 4.36IADD1936 pKa = 4.34TQNHH1940 pKa = 5.42TGLRR1944 pKa = 11.84LEE1946 pKa = 4.73AQLQIDD1952 pKa = 4.62KK1953 pKa = 10.8APQTITFTSPGMLGRR1968 pKa = 11.84DD1969 pKa = 3.64AGTVALDD1976 pKa = 3.33VRR1978 pKa = 11.84SDD1980 pKa = 3.24SGLPVSLSVDD1990 pKa = 3.27DD1991 pKa = 5.43DD1992 pKa = 3.78MVGRR1996 pKa = 11.84IEE1998 pKa = 4.17GLNLEE2003 pKa = 4.29ILRR2006 pKa = 11.84LGTVTVTAIQPGNGNYY2022 pKa = 8.47EE2023 pKa = 3.79AAEE2026 pKa = 3.97PVSFSVRR2033 pKa = 11.84VANDD2037 pKa = 2.79ATAAVPIRR2045 pKa = 11.84VHH2047 pKa = 5.9QAVSPNGDD2055 pKa = 3.81GINDD2059 pKa = 3.82FLMMEE2064 pKa = 5.35GILDD2068 pKa = 3.73YY2069 pKa = 11.02PEE2071 pKa = 4.26NKK2073 pKa = 8.16VTIFDD2078 pKa = 3.82KK2079 pKa = 10.61SGKK2082 pKa = 9.83VIDD2085 pKa = 5.03VIEE2088 pKa = 4.52SYY2090 pKa = 11.44NNRR2093 pKa = 11.84DD2094 pKa = 3.13RR2095 pKa = 11.84VFTGQFVNDD2104 pKa = 3.69GTYY2107 pKa = 10.64YY2108 pKa = 10.88YY2109 pKa = 10.78YY2110 pKa = 10.15IDD2112 pKa = 3.96VKK2114 pKa = 11.23DD2115 pKa = 3.51GGVWKK2120 pKa = 10.22RR2121 pKa = 11.84EE2122 pKa = 3.58KK2123 pKa = 11.03GFFVVKK2129 pKa = 10.55RR2130 pKa = 11.84SVNN2133 pKa = 3.26
MM1 pKa = 7.48KK2 pKa = 10.11KK3 pKa = 9.56IRR5 pKa = 11.84YY6 pKa = 8.81ILISLHH12 pKa = 6.8LVLLLLGGISSLHH25 pKa = 5.58AQCVDD30 pKa = 3.24NTGLIAFTGYY40 pKa = 10.55QIADD44 pKa = 3.28AGNPQGQSDD53 pKa = 3.92RR54 pKa = 11.84FSFVTLDD61 pKa = 3.9SIPPNTPIRR70 pKa = 11.84FTDD73 pKa = 3.09RR74 pKa = 11.84GFTGGSSFQDD84 pKa = 3.31FGNALTDD91 pKa = 4.65GDD93 pKa = 4.85FSWSDD98 pKa = 3.27ASGISPGTHH107 pKa = 4.49IVVTLGSGTATSTIGSITGYY127 pKa = 10.4LGNFNLGLSGDD138 pKa = 3.77QIFAYY143 pKa = 8.62TASPSFKK150 pKa = 9.61IHH152 pKa = 6.73AAMLINKK159 pKa = 7.29SAWDD163 pKa = 3.76TEE165 pKa = 4.05MLEE168 pKa = 4.23LGNPIVTSSKK178 pKa = 10.78SLNPAIAPVRR188 pKa = 11.84QSITVYY194 pKa = 10.96NSTDD198 pKa = 3.23DD199 pKa = 4.07AFQAKK204 pKa = 9.85LNPITLTGSRR214 pKa = 11.84TALRR218 pKa = 11.84NQLFAGIFNIANSLNTDD235 pKa = 3.19AVNLSVSPLTWNASVPTYY253 pKa = 10.74TPISLNNKK261 pKa = 8.59ILSSTVTDD269 pKa = 2.99ACYY272 pKa = 10.24IQWQSSIDD280 pKa = 3.88GINFSNINDD289 pKa = 3.56GTAFSGTKK297 pKa = 8.68TASLTINTPLVADD310 pKa = 3.77TWFRR314 pKa = 11.84MHH316 pKa = 6.74LTGAAYY322 pKa = 8.48TVTDD326 pKa = 5.33AIKK329 pKa = 9.35LTLPAVTAHH338 pKa = 6.77PLNRR342 pKa = 11.84SICVGEE348 pKa = 3.94NTSFSVTATGTDD360 pKa = 3.66LQYY363 pKa = 11.02QWQEE367 pKa = 3.73DD368 pKa = 3.85RR369 pKa = 11.84GSGFTDD375 pKa = 3.89LSDD378 pKa = 3.35GGVYY382 pKa = 10.41SGVTTSTLNISGATAGMDD400 pKa = 3.95GYY402 pKa = 10.37QYY404 pKa = 10.81RR405 pKa = 11.84VRR407 pKa = 11.84VYY409 pKa = 10.91SSIANPVTSNSATLSVSNIQATISKK434 pKa = 9.15TDD436 pKa = 3.21VGCFGEE442 pKa = 4.44ASGSATVSNVMGGIGSYY459 pKa = 8.9TYY461 pKa = 10.33SWVRR465 pKa = 11.84QGTTDD470 pKa = 3.92IISDD474 pKa = 3.85TEE476 pKa = 4.23TASGLTEE483 pKa = 3.7GTYY486 pKa = 10.1TVTIRR491 pKa = 11.84DD492 pKa = 3.72AIGCVKK498 pKa = 9.68TLDD501 pKa = 4.03VIIMQPSALTASTTQTNVSCFNGSNGTATVSVSGGTGPYY540 pKa = 8.7TYY542 pKa = 10.73SWLPSGGTSQTATGLSAGTYY562 pKa = 7.78TVTVTDD568 pKa = 4.67ANSCQITRR576 pKa = 11.84SFTITHH582 pKa = 7.15PPALMASDD590 pKa = 3.96GGKK593 pKa = 8.73TNVSCNGGSNGTATVNVVGGTGAYY617 pKa = 8.11TYY619 pKa = 9.18TYY621 pKa = 9.87SWAPFGGTSQTATGLSAGLYY641 pKa = 8.09TVTVTDD647 pKa = 4.59ANGCQTTQNFEE658 pKa = 3.77ISEE661 pKa = 4.22PTSAVVITTSDD672 pKa = 2.93IMHH675 pKa = 6.66VSTVGGSDD683 pKa = 3.21GSATVDD689 pKa = 3.36VMGGTAPYY697 pKa = 8.8TYY699 pKa = 10.06SWSPTGGTGATATGLSAGVYY719 pKa = 8.4TVTVTDD725 pKa = 5.15ANTCTYY731 pKa = 9.51SHH733 pKa = 6.68NVTIEE738 pKa = 3.91QQAVNQAPVISNLNGDD754 pKa = 3.63AVTFTEE760 pKa = 5.2DD761 pKa = 3.25GPAVLLDD768 pKa = 3.45VFGNATVSDD777 pKa = 4.37PDD779 pKa = 3.74SPDD782 pKa = 3.38FNEE785 pKa = 4.23GNLTVSIVTNGVAAEE800 pKa = 4.0DD801 pKa = 3.6RR802 pKa = 11.84LGIRR806 pKa = 11.84HH807 pKa = 6.3EE808 pKa = 4.5GSGPGLIDD816 pKa = 3.31VAGGDD821 pKa = 3.8VYY823 pKa = 11.64YY824 pKa = 10.75GGTPIGSLTGGIAGQDD840 pKa = 3.25LVVTFLPNSTPAAAQALIRR859 pKa = 11.84NLTYY863 pKa = 11.39SNVNTGNPSSAPRR876 pKa = 11.84TVYY879 pKa = 8.89ITLSDD884 pKa = 3.78GDD886 pKa = 4.25GGTSAVQAVTVSIKK900 pKa = 9.55TVNDD904 pKa = 3.28APTISVPVQINVTEE918 pKa = 4.25DD919 pKa = 3.35VPVALTGITFYY930 pKa = 11.22DD931 pKa = 3.45VDD933 pKa = 3.92NTDD936 pKa = 2.92VTVTFRR942 pKa = 11.84TTIAGAGSGTLTATSTADD960 pKa = 3.33VTVSGSGTNEE970 pKa = 3.43LTMAGPWASINNLIAANGLVFLTDD994 pKa = 3.32QDD996 pKa = 3.8NVNTVFLNIQVSDD1009 pKa = 3.83GALADD1014 pKa = 3.73STATSLIVAAVNDD1027 pKa = 4.09APQISVPLLQTVLQDD1042 pKa = 3.25ATLTFNAANGNPISISDD1059 pKa = 3.58VDD1061 pKa = 4.34AGTNPIKK1068 pKa = 11.01LNLTTTRR1075 pKa = 11.84GLLSLGSTNGLSFDD1089 pKa = 4.28IGNGTADD1096 pKa = 3.18NTMEE1100 pKa = 4.25FTGTLADD1107 pKa = 4.31INAALNGLAFEE1118 pKa = 4.55PTGGYY1123 pKa = 9.51YY1124 pKa = 10.57GPAAINITADD1134 pKa = 3.51DD1135 pKa = 3.77QGATGAGGPKK1145 pKa = 9.87SVTRR1149 pKa = 11.84EE1150 pKa = 3.47IPITVEE1156 pKa = 3.45PLNPKK1161 pKa = 7.98VTSVSASTADD1171 pKa = 3.65GLYY1174 pKa = 10.64KK1175 pKa = 10.38ADD1177 pKa = 4.61DD1178 pKa = 4.59VITLTVTFNQNVVVNTNGGDD1198 pKa = 3.57TPTLLLEE1205 pKa = 4.11TGIIDD1210 pKa = 4.16RR1211 pKa = 11.84SAIYY1215 pKa = 9.77MGGSGSNTLTFIYY1228 pKa = 9.41TVQPGDD1234 pKa = 3.3VSADD1238 pKa = 3.35LDD1240 pKa = 4.13YY1241 pKa = 11.8VSISALSLNGATILSSYY1258 pKa = 10.51NGANAVLTLPTVGGASSIAGQKK1280 pKa = 10.51NIVIDD1285 pKa = 4.54GIVPVVNAVAVPANAYY1301 pKa = 9.63YY1302 pKa = 10.94GSGDD1306 pKa = 3.3QLYY1309 pKa = 7.98FTVEE1313 pKa = 3.92FSEE1316 pKa = 4.38AVAVNTTGGSPALSLTIGSQTVSAQYY1342 pKa = 9.05LTEE1345 pKa = 4.36SSTTTDD1351 pKa = 3.19LLFRR1355 pKa = 11.84YY1356 pKa = 9.27SVAAGMQDD1364 pKa = 2.72HH1365 pKa = 7.46DD1366 pKa = 4.9GIEE1369 pKa = 4.07IGTLSLNGATIRR1381 pKa = 11.84DD1382 pKa = 3.65AAGNDD1387 pKa = 3.48AVLTLNNVGATTHH1400 pKa = 5.63VKK1402 pKa = 10.04VDD1404 pKa = 3.36AKK1406 pKa = 11.08SPVVTMTINGGATHH1420 pKa = 6.53TNDD1423 pKa = 3.65RR1424 pKa = 11.84NVTLTLTTADD1434 pKa = 3.33TDD1436 pKa = 3.87VATMSFMNSPIFFPGSGASQPEE1458 pKa = 4.03PFAPTKK1464 pKa = 10.3AWQLTAGDD1472 pKa = 3.69GMKK1475 pKa = 9.97FVSGIVIDD1483 pKa = 4.04GAGNLTSVLAVITLDD1498 pKa = 3.1QTAPIVTGVDD1508 pKa = 2.82EE1509 pKa = 4.88GEE1511 pKa = 4.48SYY1513 pKa = 9.37NTDD1516 pKa = 2.8RR1517 pKa = 11.84TITFNEE1523 pKa = 4.26GTATLNGATFISGTTVSQDD1542 pKa = 2.77GAYY1545 pKa = 9.12TLIVTDD1551 pKa = 3.64EE1552 pKa = 4.75AGNSTTLSFAIDD1564 pKa = 3.35QTAPAVPAGLTATPDD1579 pKa = 3.14AGNINLQWTANQEE1592 pKa = 3.93ADD1594 pKa = 3.18LAAYY1598 pKa = 10.09RR1599 pKa = 11.84LYY1601 pKa = 11.0SGTAADD1607 pKa = 4.0QLSVLADD1614 pKa = 3.65IPAGTTTYY1622 pKa = 9.45THH1624 pKa = 6.96GPLTMGQRR1632 pKa = 11.84YY1633 pKa = 9.05YY1634 pKa = 11.24YY1635 pKa = 10.52AITAIDD1641 pKa = 3.5ALGNEE1646 pKa = 4.75SAQSVVVDD1654 pKa = 5.08ALPQDD1659 pKa = 3.76TQTITFGTLNDD1670 pKa = 3.75MTYY1673 pKa = 11.38GDD1675 pKa = 4.77APFALTATTTSGLTVAYY1692 pKa = 9.92SSSDD1696 pKa = 3.26NSIASIAGDD1705 pKa = 3.51VLTIHH1710 pKa = 6.18QAGDD1714 pKa = 3.54VVITASQNGNNAFLAATPVTQALHH1738 pKa = 5.9INKK1741 pKa = 9.57ADD1743 pKa = 3.68FAGITFDD1750 pKa = 5.28DD1751 pKa = 3.86KK1752 pKa = 11.26TVTYY1756 pKa = 10.19DD1757 pKa = 3.76GSAHH1761 pKa = 6.8SIFVSNAPTDD1771 pKa = 3.39ATVIYY1776 pKa = 9.97TNNNQINAGTYY1787 pKa = 7.96TVMATVSQANYY1798 pKa = 10.5NDD1800 pKa = 4.14LPLTAEE1806 pKa = 4.12LTIEE1810 pKa = 4.16KK1811 pKa = 10.35ADD1813 pKa = 3.72FAGITFNDD1821 pKa = 3.25ATVTYY1826 pKa = 9.67DD1827 pKa = 3.8GSAHH1831 pKa = 6.51SIAVANTPTGATITYY1846 pKa = 9.54VGNAQINAGTYY1857 pKa = 6.69TVRR1860 pKa = 11.84ATVSQANYY1868 pKa = 10.1NDD1870 pKa = 3.87LVLTAEE1876 pKa = 4.31LEE1878 pKa = 4.07ITKK1881 pKa = 10.97AMLTDD1886 pKa = 3.23IALRR1890 pKa = 11.84NNSFVYY1896 pKa = 10.57DD1897 pKa = 4.04GNAKK1901 pKa = 10.08SLEE1904 pKa = 3.95ISGILPTGVQVIYY1917 pKa = 10.17TGNNQVNAGMYY1928 pKa = 8.74TVVAEE1933 pKa = 4.36IADD1936 pKa = 4.34TQNHH1940 pKa = 5.42TGLRR1944 pKa = 11.84LEE1946 pKa = 4.73AQLQIDD1952 pKa = 4.62KK1953 pKa = 10.8APQTITFTSPGMLGRR1968 pKa = 11.84DD1969 pKa = 3.64AGTVALDD1976 pKa = 3.33VRR1978 pKa = 11.84SDD1980 pKa = 3.24SGLPVSLSVDD1990 pKa = 3.27DD1991 pKa = 5.43DD1992 pKa = 3.78MVGRR1996 pKa = 11.84IEE1998 pKa = 4.17GLNLEE2003 pKa = 4.29ILRR2006 pKa = 11.84LGTVTVTAIQPGNGNYY2022 pKa = 8.47EE2023 pKa = 3.79AAEE2026 pKa = 3.97PVSFSVRR2033 pKa = 11.84VANDD2037 pKa = 2.79ATAAVPIRR2045 pKa = 11.84VHH2047 pKa = 5.9QAVSPNGDD2055 pKa = 3.81GINDD2059 pKa = 3.82FLMMEE2064 pKa = 5.35GILDD2068 pKa = 3.73YY2069 pKa = 11.02PEE2071 pKa = 4.26NKK2073 pKa = 8.16VTIFDD2078 pKa = 3.82KK2079 pKa = 10.61SGKK2082 pKa = 9.83VIDD2085 pKa = 5.03VIEE2088 pKa = 4.52SYY2090 pKa = 11.44NNRR2093 pKa = 11.84DD2094 pKa = 3.13RR2095 pKa = 11.84VFTGQFVNDD2104 pKa = 3.69GTYY2107 pKa = 10.64YY2108 pKa = 10.88YY2109 pKa = 10.78YY2110 pKa = 10.15IDD2112 pKa = 3.96VKK2114 pKa = 11.23DD2115 pKa = 3.51GGVWKK2120 pKa = 10.22RR2121 pKa = 11.84EE2122 pKa = 3.58KK2123 pKa = 11.03GFFVVKK2129 pKa = 10.55RR2130 pKa = 11.84SVNN2133 pKa = 3.26
Molecular weight: 220.57 kDa
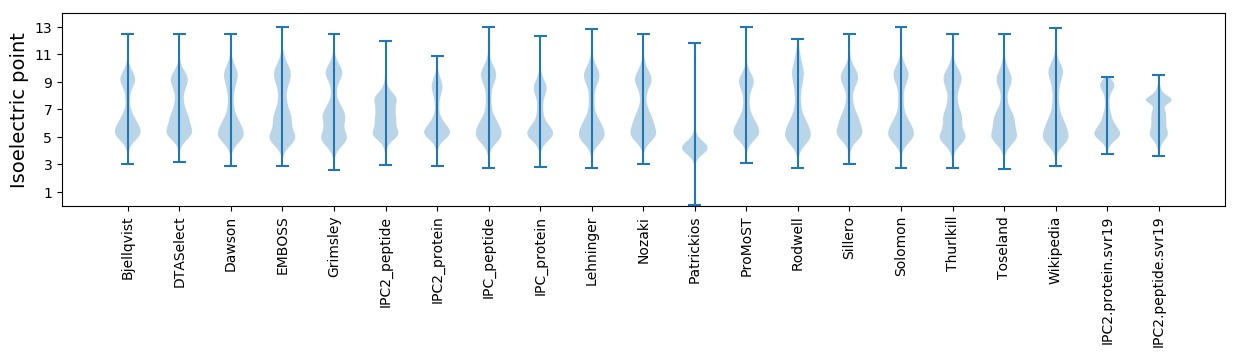
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I6NV61|A0A1I6NV61_9SPHI De-hypoxanthine futalosine cyclase OS=Sphingobacterium wenxiniae OX=683125 GN=SAMN05660206_101117 PE=4 SV=1
MM1 pKa = 7.29YY2 pKa = 10.36NGSLVMSFRR11 pKa = 11.84PLVMNIGSLVMSIRR25 pKa = 11.84SLVMSIRR32 pKa = 11.84PLVMSIRR39 pKa = 11.84PLVMSFRR46 pKa = 11.84LLVMSFRR53 pKa = 11.84PLVMSIRR60 pKa = 11.84SLVMSIRR67 pKa = 11.84SLVMSIRR74 pKa = 11.84LLVMSIRR81 pKa = 11.84PLVMSFRR88 pKa = 11.84PLVMNIGSLVMNIRR102 pKa = 11.84PLLMHH107 pKa = 6.63IGKK110 pKa = 8.87WLLTEE115 pKa = 4.15KK116 pKa = 10.9
MM1 pKa = 7.29YY2 pKa = 10.36NGSLVMSFRR11 pKa = 11.84PLVMNIGSLVMSIRR25 pKa = 11.84SLVMSIRR32 pKa = 11.84PLVMSIRR39 pKa = 11.84PLVMSFRR46 pKa = 11.84LLVMSFRR53 pKa = 11.84PLVMSIRR60 pKa = 11.84SLVMSIRR67 pKa = 11.84SLVMSIRR74 pKa = 11.84LLVMSIRR81 pKa = 11.84PLVMSFRR88 pKa = 11.84PLVMNIGSLVMNIRR102 pKa = 11.84PLLMHH107 pKa = 6.63IGKK110 pKa = 8.87WLLTEE115 pKa = 4.15KK116 pKa = 10.9
Molecular weight: 13.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1141595 |
14 |
3684 |
334.6 |
37.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.082 ± 0.043 | 0.739 ± 0.014 |
5.482 ± 0.029 | 6.483 ± 0.045 |
4.912 ± 0.032 | 6.709 ± 0.043 |
1.982 ± 0.022 | 7.266 ± 0.038 |
6.704 ± 0.045 | 9.51 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.32 ± 0.019 | 5.317 ± 0.04 |
3.565 ± 0.021 | 4.019 ± 0.027 |
4.232 ± 0.028 | 6.201 ± 0.03 |
5.581 ± 0.043 | 6.529 ± 0.035 |
1.155 ± 0.017 | 4.213 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
