
Tobacco vein distorting virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus
Average proteome isoelectric point is 7.7
Get precalculated fractions of proteins
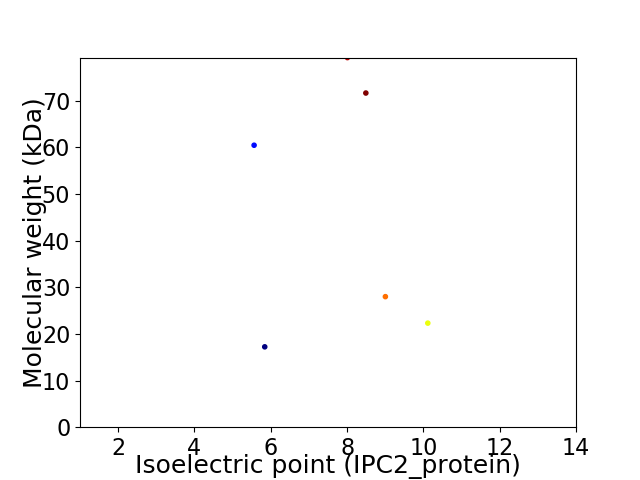
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B2BJG5|B2BJG5_9LUTE Readthrough protein OS=Tobacco vein distorting virus OX=192203 GN=RT PE=3 SV=1
MM1 pKa = 6.92FRR3 pKa = 11.84EE4 pKa = 4.17AFGEE8 pKa = 3.94NGGKK12 pKa = 9.14NRR14 pKa = 11.84PFCYY18 pKa = 9.56RR19 pKa = 11.84KK20 pKa = 9.86EE21 pKa = 4.28SGGSISTEE29 pKa = 3.8SHH31 pKa = 6.08EE32 pKa = 4.34EE33 pKa = 3.58ASRR36 pKa = 11.84IPXAQTTAEE45 pKa = 3.76NFRR48 pKa = 11.84RR49 pKa = 11.84YY50 pKa = 9.96FEE52 pKa = 5.39GKK54 pKa = 10.13YY55 pKa = 9.53KK56 pKa = 10.3WEE58 pKa = 4.4VPASPQEE65 pKa = 3.76IPGFEE70 pKa = 4.24EE71 pKa = 4.98CGSLPQYY78 pKa = 8.33YY79 pKa = 9.71HH80 pKa = 7.18PKK82 pKa = 9.31QIKK85 pKa = 8.64GSQWGIQLIQEE96 pKa = 4.21HH97 pKa = 7.26PEE99 pKa = 3.96LGEE102 pKa = 4.1KK103 pKa = 10.61VSGFGWPAVGPQAEE117 pKa = 4.69VTSLTLQAEE126 pKa = 4.3RR127 pKa = 11.84WLQRR131 pKa = 11.84AQSAKK136 pKa = 9.98IPSSEE141 pKa = 3.75DD142 pKa = 3.04RR143 pKa = 11.84EE144 pKa = 4.04RR145 pKa = 11.84VINKK149 pKa = 7.96TVEE152 pKa = 3.93AYY154 pKa = 11.26SNVKK158 pKa = 9.07TFGPTATRR166 pKa = 11.84GNKK169 pKa = 9.98LEE171 pKa = 3.74WRR173 pKa = 11.84QFIEE177 pKa = 4.25DD178 pKa = 3.8FKK180 pKa = 11.45SAVFSLEE187 pKa = 3.4LDD189 pKa = 3.35AGIGVPYY196 pKa = 9.85IAYY199 pKa = 9.28GRR201 pKa = 11.84PTHH204 pKa = 6.38KK205 pKa = 10.41GWVEE209 pKa = 3.94DD210 pKa = 4.03PKK212 pKa = 11.09LLPVLARR219 pKa = 11.84LTFNRR224 pKa = 11.84LQKK227 pKa = 9.49MLEE230 pKa = 4.16VEE232 pKa = 4.18SSEE235 pKa = 3.98MSAEE239 pKa = 4.07EE240 pKa = 3.87LVQAGLCDD248 pKa = 4.92PIRR251 pKa = 11.84TFVKK255 pKa = 10.42RR256 pKa = 11.84EE257 pKa = 3.58PHH259 pKa = 5.86KK260 pKa = 10.47QSKK263 pKa = 10.11LDD265 pKa = 3.38EE266 pKa = 4.27GRR268 pKa = 11.84YY269 pKa = 8.96RR270 pKa = 11.84LIMSVSLVDD279 pKa = 3.45QLVARR284 pKa = 11.84VLFQNQNKK292 pKa = 9.66RR293 pKa = 11.84EE294 pKa = 3.96IALWRR299 pKa = 11.84ANPSKK304 pKa = 10.61PGFGLSTDD312 pKa = 3.63EE313 pKa = 4.0QVLEE317 pKa = 4.39FVQALAAQVEE327 pKa = 4.75VPPEE331 pKa = 3.73EE332 pKa = 5.59VITSWEE338 pKa = 4.24KK339 pKa = 10.58YY340 pKa = 9.87LVPTDD345 pKa = 3.67CSGFDD350 pKa = 3.21WSVAEE355 pKa = 4.57WMLHH359 pKa = 5.89DD360 pKa = 5.83DD361 pKa = 3.43MVVRR365 pKa = 11.84NKK367 pKa = 9.41LTLDD371 pKa = 4.03LNPTTEE377 pKa = 4.11KK378 pKa = 10.92LRR380 pKa = 11.84FAWLKK385 pKa = 10.59CISNSVLCLSDD396 pKa = 3.31GTLLXQRR403 pKa = 11.84VPGVQKK409 pKa = 10.45SGSYY413 pKa = 8.25NTSSSNSRR421 pKa = 11.84IRR423 pKa = 11.84VMAAYY428 pKa = 9.95HH429 pKa = 6.59CGADD433 pKa = 2.88WAMAMGDD440 pKa = 3.72DD441 pKa = 4.39ALEE444 pKa = 4.26SVNTNLEE451 pKa = 4.32VYY453 pKa = 10.11KK454 pKa = 10.98SLGFKK459 pKa = 10.72VEE461 pKa = 3.93VSGQLEE467 pKa = 4.4FCSHH471 pKa = 6.9IFRR474 pKa = 11.84APDD477 pKa = 2.96LALPVNEE484 pKa = 4.57RR485 pKa = 11.84KK486 pKa = 8.59MLYY489 pKa = 10.45KK490 pKa = 10.76LIFGYY495 pKa = 10.83NPGSGSLEE503 pKa = 4.12VISNYY508 pKa = 8.13XAACASVLNEE518 pKa = 4.41LRR520 pKa = 11.84HH521 pKa = 6.43DD522 pKa = 4.37PDD524 pKa = 4.09SVALLXXWLVHH535 pKa = 6.39PVLPQNDD542 pKa = 3.36
MM1 pKa = 6.92FRR3 pKa = 11.84EE4 pKa = 4.17AFGEE8 pKa = 3.94NGGKK12 pKa = 9.14NRR14 pKa = 11.84PFCYY18 pKa = 9.56RR19 pKa = 11.84KK20 pKa = 9.86EE21 pKa = 4.28SGGSISTEE29 pKa = 3.8SHH31 pKa = 6.08EE32 pKa = 4.34EE33 pKa = 3.58ASRR36 pKa = 11.84IPXAQTTAEE45 pKa = 3.76NFRR48 pKa = 11.84RR49 pKa = 11.84YY50 pKa = 9.96FEE52 pKa = 5.39GKK54 pKa = 10.13YY55 pKa = 9.53KK56 pKa = 10.3WEE58 pKa = 4.4VPASPQEE65 pKa = 3.76IPGFEE70 pKa = 4.24EE71 pKa = 4.98CGSLPQYY78 pKa = 8.33YY79 pKa = 9.71HH80 pKa = 7.18PKK82 pKa = 9.31QIKK85 pKa = 8.64GSQWGIQLIQEE96 pKa = 4.21HH97 pKa = 7.26PEE99 pKa = 3.96LGEE102 pKa = 4.1KK103 pKa = 10.61VSGFGWPAVGPQAEE117 pKa = 4.69VTSLTLQAEE126 pKa = 4.3RR127 pKa = 11.84WLQRR131 pKa = 11.84AQSAKK136 pKa = 9.98IPSSEE141 pKa = 3.75DD142 pKa = 3.04RR143 pKa = 11.84EE144 pKa = 4.04RR145 pKa = 11.84VINKK149 pKa = 7.96TVEE152 pKa = 3.93AYY154 pKa = 11.26SNVKK158 pKa = 9.07TFGPTATRR166 pKa = 11.84GNKK169 pKa = 9.98LEE171 pKa = 3.74WRR173 pKa = 11.84QFIEE177 pKa = 4.25DD178 pKa = 3.8FKK180 pKa = 11.45SAVFSLEE187 pKa = 3.4LDD189 pKa = 3.35AGIGVPYY196 pKa = 9.85IAYY199 pKa = 9.28GRR201 pKa = 11.84PTHH204 pKa = 6.38KK205 pKa = 10.41GWVEE209 pKa = 3.94DD210 pKa = 4.03PKK212 pKa = 11.09LLPVLARR219 pKa = 11.84LTFNRR224 pKa = 11.84LQKK227 pKa = 9.49MLEE230 pKa = 4.16VEE232 pKa = 4.18SSEE235 pKa = 3.98MSAEE239 pKa = 4.07EE240 pKa = 3.87LVQAGLCDD248 pKa = 4.92PIRR251 pKa = 11.84TFVKK255 pKa = 10.42RR256 pKa = 11.84EE257 pKa = 3.58PHH259 pKa = 5.86KK260 pKa = 10.47QSKK263 pKa = 10.11LDD265 pKa = 3.38EE266 pKa = 4.27GRR268 pKa = 11.84YY269 pKa = 8.96RR270 pKa = 11.84LIMSVSLVDD279 pKa = 3.45QLVARR284 pKa = 11.84VLFQNQNKK292 pKa = 9.66RR293 pKa = 11.84EE294 pKa = 3.96IALWRR299 pKa = 11.84ANPSKK304 pKa = 10.61PGFGLSTDD312 pKa = 3.63EE313 pKa = 4.0QVLEE317 pKa = 4.39FVQALAAQVEE327 pKa = 4.75VPPEE331 pKa = 3.73EE332 pKa = 5.59VITSWEE338 pKa = 4.24KK339 pKa = 10.58YY340 pKa = 9.87LVPTDD345 pKa = 3.67CSGFDD350 pKa = 3.21WSVAEE355 pKa = 4.57WMLHH359 pKa = 5.89DD360 pKa = 5.83DD361 pKa = 3.43MVVRR365 pKa = 11.84NKK367 pKa = 9.41LTLDD371 pKa = 4.03LNPTTEE377 pKa = 4.11KK378 pKa = 10.92LRR380 pKa = 11.84FAWLKK385 pKa = 10.59CISNSVLCLSDD396 pKa = 3.31GTLLXQRR403 pKa = 11.84VPGVQKK409 pKa = 10.45SGSYY413 pKa = 8.25NTSSSNSRR421 pKa = 11.84IRR423 pKa = 11.84VMAAYY428 pKa = 9.95HH429 pKa = 6.59CGADD433 pKa = 2.88WAMAMGDD440 pKa = 3.72DD441 pKa = 4.39ALEE444 pKa = 4.26SVNTNLEE451 pKa = 4.32VYY453 pKa = 10.11KK454 pKa = 10.98SLGFKK459 pKa = 10.72VEE461 pKa = 3.93VSGQLEE467 pKa = 4.4FCSHH471 pKa = 6.9IFRR474 pKa = 11.84APDD477 pKa = 2.96LALPVNEE484 pKa = 4.57RR485 pKa = 11.84KK486 pKa = 8.59MLYY489 pKa = 10.45KK490 pKa = 10.76LIFGYY495 pKa = 10.83NPGSGSLEE503 pKa = 4.12VISNYY508 pKa = 8.13XAACASVLNEE518 pKa = 4.41LRR520 pKa = 11.84HH521 pKa = 6.43DD522 pKa = 4.37PDD524 pKa = 4.09SVALLXXWLVHH535 pKa = 6.39PVLPQNDD542 pKa = 3.36
Molecular weight: 60.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B2BJG7|B2BJG7_9LUTE Movement protein OS=Tobacco vein distorting virus OX=192203 GN=MP PE=3 SV=1
MM1 pKa = 6.74NTGGARR7 pKa = 11.84SNNGNGGSRR16 pKa = 11.84VSRR19 pKa = 11.84PRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84ARR25 pKa = 11.84SVRR28 pKa = 11.84PVVVVAPPRR37 pKa = 11.84GARR40 pKa = 11.84XRR42 pKa = 11.84TRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84NGGRR51 pKa = 11.84NRR53 pKa = 11.84RR54 pKa = 11.84GRR56 pKa = 11.84NGVGGRR62 pKa = 11.84SSNSEE67 pKa = 3.69TFIFNKK73 pKa = 10.42DD74 pKa = 3.59SIKK77 pKa = 10.81DD78 pKa = 3.66SSSGSITFGPSLSEE92 pKa = 4.12SVALSGGVLKK102 pKa = 10.54AYY104 pKa = 10.22HH105 pKa = 6.68EE106 pKa = 4.5YY107 pKa = 10.51KK108 pKa = 9.32ITMVNIRR115 pKa = 11.84FISEE119 pKa = 4.02SSSTAEE125 pKa = 3.6GSIAYY130 pKa = 9.39EE131 pKa = 4.08LDD133 pKa = 3.42PHH135 pKa = 6.93CKK137 pKa = 10.08LSSLQSTLRR146 pKa = 11.84KK147 pKa = 9.77FPVTKK152 pKa = 10.36GGQATFRR159 pKa = 11.84AAQINGVEE167 pKa = 4.07WHH169 pKa = 7.1DD170 pKa = 3.55TTEE173 pKa = 3.94DD174 pKa = 3.47QFRR177 pKa = 11.84LLYY180 pKa = 10.2KK181 pKa = 10.92GNGTKK186 pKa = 10.41GVAAGFFQIRR196 pKa = 11.84YY197 pKa = 6.05TVQLHH202 pKa = 5.2NPKK205 pKa = 10.54
MM1 pKa = 6.74NTGGARR7 pKa = 11.84SNNGNGGSRR16 pKa = 11.84VSRR19 pKa = 11.84PRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84ARR25 pKa = 11.84SVRR28 pKa = 11.84PVVVVAPPRR37 pKa = 11.84GARR40 pKa = 11.84XRR42 pKa = 11.84TRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84NGGRR51 pKa = 11.84NRR53 pKa = 11.84RR54 pKa = 11.84GRR56 pKa = 11.84NGVGGRR62 pKa = 11.84SSNSEE67 pKa = 3.69TFIFNKK73 pKa = 10.42DD74 pKa = 3.59SIKK77 pKa = 10.81DD78 pKa = 3.66SSSGSITFGPSLSEE92 pKa = 4.12SVALSGGVLKK102 pKa = 10.54AYY104 pKa = 10.22HH105 pKa = 6.68EE106 pKa = 4.5YY107 pKa = 10.51KK108 pKa = 9.32ITMVNIRR115 pKa = 11.84FISEE119 pKa = 4.02SSSTAEE125 pKa = 3.6GSIAYY130 pKa = 9.39EE131 pKa = 4.08LDD133 pKa = 3.42PHH135 pKa = 6.93CKK137 pKa = 10.08LSSLQSTLRR146 pKa = 11.84KK147 pKa = 9.77FPVTKK152 pKa = 10.36GGQATFRR159 pKa = 11.84AAQINGVEE167 pKa = 4.07WHH169 pKa = 7.1DD170 pKa = 3.55TTEE173 pKa = 3.94DD174 pKa = 3.47QFRR177 pKa = 11.84LLYY180 pKa = 10.2KK181 pKa = 10.92GNGTKK186 pKa = 10.41GVAAGFFQIRR196 pKa = 11.84YY197 pKa = 6.05TVQLHH202 pKa = 5.2NPKK205 pKa = 10.54
Molecular weight: 22.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2533 |
156 |
723 |
422.2 |
46.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.711 ± 0.315 | 1.303 ± 0.282 |
3.908 ± 0.772 | 6.514 ± 0.711 |
3.829 ± 0.365 | 7.58 ± 0.721 |
2.132 ± 0.276 | 3.79 ± 0.337 |
5.527 ± 0.658 | 8.725 ± 0.91 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.856 ± 0.229 | 4.501 ± 0.413 |
6.396 ± 0.547 | 4.224 ± 0.281 |
6.79 ± 1.151 | 9.988 ± 0.605 |
5.448 ± 0.523 | 6.04 ± 0.8 |
1.54 ± 0.344 | 2.487 ± 0.362 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
