
Botrytis cinerea mitovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 8.71
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
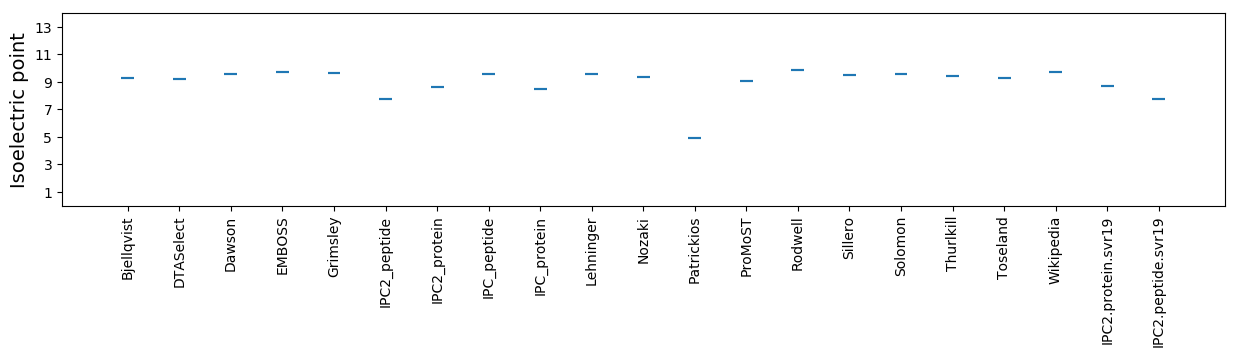
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A5YRF7|A5YRF7_9VIRU RNA-dependent RNA polymerase OS=Botrytis cinerea mitovirus 1 OX=444193 GN=RDRP PE=4 SV=3
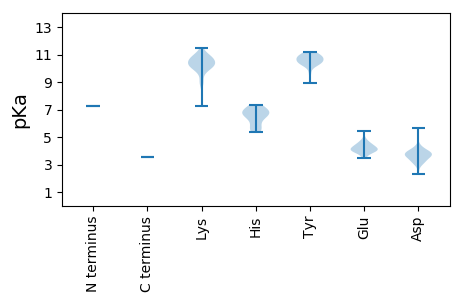
MM1 pKa = 7.27KK2 pKa = 10.2HH3 pKa = 6.41LFRR6 pKa = 11.84IKK8 pKa = 10.14RR9 pKa = 11.84KK10 pKa = 9.69AVIKK14 pKa = 10.78NSTTRR19 pKa = 11.84LPDD22 pKa = 3.96HH23 pKa = 6.8LWLGYY28 pKa = 10.63VKK30 pKa = 10.7VLVWLYY36 pKa = 9.65DD37 pKa = 3.18IPSAHH42 pKa = 5.68RR43 pKa = 11.84TYY45 pKa = 10.8FFEE48 pKa = 4.47LLNRR52 pKa = 11.84IQSLRR57 pKa = 11.84RR58 pKa = 11.84NSGDD62 pKa = 2.81TWTVGYY68 pKa = 10.28LKK70 pKa = 10.26EE71 pKa = 4.06SHH73 pKa = 6.64RR74 pKa = 11.84LSSHH78 pKa = 5.36WLSGNPDD85 pKa = 3.21VCSGSIRR92 pKa = 11.84VSVRR96 pKa = 11.84GLPLIIPGRR105 pKa = 11.84LRR107 pKa = 11.84NVIYY111 pKa = 10.55SRR113 pKa = 11.84NNINITRR120 pKa = 11.84ALLTLLSTFRR130 pKa = 11.84VMSATPKK137 pKa = 10.49LKK139 pKa = 10.63LSTITDD145 pKa = 3.73PFNGVNEE152 pKa = 4.34TLPSLEE158 pKa = 3.91VSRR161 pKa = 11.84ALKK164 pKa = 10.74NILQGNKK171 pKa = 7.26FHH173 pKa = 7.32FKK175 pKa = 10.76KK176 pKa = 10.62EE177 pKa = 4.33DD178 pKa = 3.09IKK180 pKa = 11.45DD181 pKa = 3.69PYY183 pKa = 10.6SVNRR187 pKa = 11.84LMDD190 pKa = 3.2ITKK193 pKa = 10.1ADD195 pKa = 3.87PNGGVSLRR203 pKa = 11.84NVIIDD208 pKa = 3.97AYY210 pKa = 10.95ALMKK214 pKa = 10.0HH215 pKa = 5.83PKK217 pKa = 9.69VYY219 pKa = 10.89ASLLEE224 pKa = 4.2LSNVMSPYY232 pKa = 10.47LGSLLRR238 pKa = 11.84NQTTIIRR245 pKa = 11.84AIIDD249 pKa = 3.51YY250 pKa = 10.87DD251 pKa = 4.11PIEE254 pKa = 4.1WEE256 pKa = 4.05RR257 pKa = 11.84IKK259 pKa = 11.07PFMAPIYY266 pKa = 10.46QNIKK270 pKa = 10.69DD271 pKa = 3.84RR272 pKa = 11.84TLKK275 pKa = 10.6LVPDD279 pKa = 4.32LSCIGKK285 pKa = 10.29LSTKK289 pKa = 10.2EE290 pKa = 3.51EE291 pKa = 4.03AAGKK295 pKa = 9.74VRR297 pKa = 11.84VFAMVDD303 pKa = 2.32IWTQSILNPLHH314 pKa = 6.71KK315 pKa = 10.3KK316 pKa = 8.81IFSIIRR322 pKa = 11.84EE323 pKa = 4.43LPTDD327 pKa = 3.6GTFDD331 pKa = 3.33QLKK334 pKa = 10.04PLDD337 pKa = 4.18RR338 pKa = 11.84LHH340 pKa = 6.86EE341 pKa = 4.63LSTQDD346 pKa = 3.61RR347 pKa = 11.84FSFDD351 pKa = 3.18LSAATDD357 pKa = 3.93RR358 pKa = 11.84LPLTLQKK365 pKa = 10.86DD366 pKa = 3.44ILTLLVSPSFAEE378 pKa = 3.88AWGTALVGRR387 pKa = 11.84PYY389 pKa = 10.38KK390 pKa = 10.01WKK392 pKa = 10.49FGTTEE397 pKa = 4.12DD398 pKa = 3.49EE399 pKa = 4.12LMYY402 pKa = 10.94SVGQPMGALSSWGMLALTHH421 pKa = 5.49HH422 pKa = 7.14TIVQVAASRR431 pKa = 11.84AGYY434 pKa = 10.29KK435 pKa = 10.44DD436 pKa = 4.38LFLDD440 pKa = 3.91YY441 pKa = 11.17ALLGDD446 pKa = 5.67DD447 pKa = 3.56ICIANKK453 pKa = 10.14AVADD457 pKa = 3.94NYY459 pKa = 10.88LLIMRR464 pKa = 11.84DD465 pKa = 3.3LGVEE469 pKa = 3.92INLSKK474 pKa = 11.05SLISSTGVVEE484 pKa = 4.74FAKK487 pKa = 10.13RR488 pKa = 11.84WRR490 pKa = 11.84VGQTDD495 pKa = 3.66VSPASPALITRR506 pKa = 11.84LLSNMNYY513 pKa = 10.27LPVLILDD520 pKa = 4.13LVNRR524 pKa = 11.84GVKK527 pKa = 9.54TIQNSEE533 pKa = 4.18KK534 pKa = 10.53LLSLSDD540 pKa = 3.79SLHH543 pKa = 6.42KK544 pKa = 10.14IKK546 pKa = 10.79KK547 pKa = 9.98SITFSLIPYY556 pKa = 8.96YY557 pKa = 10.63DD558 pKa = 3.74SEE560 pKa = 4.35FAKK563 pKa = 10.79CLLPLKK569 pKa = 10.65GKK571 pKa = 10.21DD572 pKa = 3.05KK573 pKa = 11.26SFSQKK578 pKa = 10.14EE579 pKa = 3.86LFALYY584 pKa = 10.42VHH586 pKa = 6.66TDD588 pKa = 3.85RR589 pKa = 11.84IFNKK593 pKa = 10.27LGLLEE598 pKa = 3.8FHH600 pKa = 7.02KK601 pKa = 11.06AKK603 pKa = 10.99DD604 pKa = 3.59GDD606 pKa = 3.92QEE608 pKa = 4.12NSKK611 pKa = 10.69LSQSYY616 pKa = 10.25LYY618 pKa = 10.77NILDD622 pKa = 3.72NSKK625 pKa = 10.83LPLPAITSQIDD636 pKa = 3.57NILIAQMKK644 pKa = 8.92TITNVPTRR652 pKa = 11.84NWLNNRR658 pKa = 11.84DD659 pKa = 3.55INLLTFEE666 pKa = 4.86CWHH669 pKa = 6.87SYY671 pKa = 10.9LIDD674 pKa = 3.76TLSEE678 pKa = 3.88LSSISKK684 pKa = 9.02VVPDD688 pKa = 3.14YY689 pKa = 11.18TIIKK693 pKa = 10.41DD694 pKa = 3.67NEE696 pKa = 4.19KK697 pKa = 10.41PDD699 pKa = 3.83NLEE702 pKa = 3.45MRR704 pKa = 11.84RR705 pKa = 11.84NLLFMQEE712 pKa = 4.53LIKK715 pKa = 10.59DD716 pKa = 3.99LKK718 pKa = 10.13KK719 pKa = 9.04WNPGIFEE726 pKa = 5.45IIPEE730 pKa = 4.53SILKK734 pKa = 10.26EE735 pKa = 4.12EE736 pKa = 4.37EE737 pKa = 4.03VV738 pKa = 3.56
MM1 pKa = 7.27KK2 pKa = 10.2HH3 pKa = 6.41LFRR6 pKa = 11.84IKK8 pKa = 10.14RR9 pKa = 11.84KK10 pKa = 9.69AVIKK14 pKa = 10.78NSTTRR19 pKa = 11.84LPDD22 pKa = 3.96HH23 pKa = 6.8LWLGYY28 pKa = 10.63VKK30 pKa = 10.7VLVWLYY36 pKa = 9.65DD37 pKa = 3.18IPSAHH42 pKa = 5.68RR43 pKa = 11.84TYY45 pKa = 10.8FFEE48 pKa = 4.47LLNRR52 pKa = 11.84IQSLRR57 pKa = 11.84RR58 pKa = 11.84NSGDD62 pKa = 2.81TWTVGYY68 pKa = 10.28LKK70 pKa = 10.26EE71 pKa = 4.06SHH73 pKa = 6.64RR74 pKa = 11.84LSSHH78 pKa = 5.36WLSGNPDD85 pKa = 3.21VCSGSIRR92 pKa = 11.84VSVRR96 pKa = 11.84GLPLIIPGRR105 pKa = 11.84LRR107 pKa = 11.84NVIYY111 pKa = 10.55SRR113 pKa = 11.84NNINITRR120 pKa = 11.84ALLTLLSTFRR130 pKa = 11.84VMSATPKK137 pKa = 10.49LKK139 pKa = 10.63LSTITDD145 pKa = 3.73PFNGVNEE152 pKa = 4.34TLPSLEE158 pKa = 3.91VSRR161 pKa = 11.84ALKK164 pKa = 10.74NILQGNKK171 pKa = 7.26FHH173 pKa = 7.32FKK175 pKa = 10.76KK176 pKa = 10.62EE177 pKa = 4.33DD178 pKa = 3.09IKK180 pKa = 11.45DD181 pKa = 3.69PYY183 pKa = 10.6SVNRR187 pKa = 11.84LMDD190 pKa = 3.2ITKK193 pKa = 10.1ADD195 pKa = 3.87PNGGVSLRR203 pKa = 11.84NVIIDD208 pKa = 3.97AYY210 pKa = 10.95ALMKK214 pKa = 10.0HH215 pKa = 5.83PKK217 pKa = 9.69VYY219 pKa = 10.89ASLLEE224 pKa = 4.2LSNVMSPYY232 pKa = 10.47LGSLLRR238 pKa = 11.84NQTTIIRR245 pKa = 11.84AIIDD249 pKa = 3.51YY250 pKa = 10.87DD251 pKa = 4.11PIEE254 pKa = 4.1WEE256 pKa = 4.05RR257 pKa = 11.84IKK259 pKa = 11.07PFMAPIYY266 pKa = 10.46QNIKK270 pKa = 10.69DD271 pKa = 3.84RR272 pKa = 11.84TLKK275 pKa = 10.6LVPDD279 pKa = 4.32LSCIGKK285 pKa = 10.29LSTKK289 pKa = 10.2EE290 pKa = 3.51EE291 pKa = 4.03AAGKK295 pKa = 9.74VRR297 pKa = 11.84VFAMVDD303 pKa = 2.32IWTQSILNPLHH314 pKa = 6.71KK315 pKa = 10.3KK316 pKa = 8.81IFSIIRR322 pKa = 11.84EE323 pKa = 4.43LPTDD327 pKa = 3.6GTFDD331 pKa = 3.33QLKK334 pKa = 10.04PLDD337 pKa = 4.18RR338 pKa = 11.84LHH340 pKa = 6.86EE341 pKa = 4.63LSTQDD346 pKa = 3.61RR347 pKa = 11.84FSFDD351 pKa = 3.18LSAATDD357 pKa = 3.93RR358 pKa = 11.84LPLTLQKK365 pKa = 10.86DD366 pKa = 3.44ILTLLVSPSFAEE378 pKa = 3.88AWGTALVGRR387 pKa = 11.84PYY389 pKa = 10.38KK390 pKa = 10.01WKK392 pKa = 10.49FGTTEE397 pKa = 4.12DD398 pKa = 3.49EE399 pKa = 4.12LMYY402 pKa = 10.94SVGQPMGALSSWGMLALTHH421 pKa = 5.49HH422 pKa = 7.14TIVQVAASRR431 pKa = 11.84AGYY434 pKa = 10.29KK435 pKa = 10.44DD436 pKa = 4.38LFLDD440 pKa = 3.91YY441 pKa = 11.17ALLGDD446 pKa = 5.67DD447 pKa = 3.56ICIANKK453 pKa = 10.14AVADD457 pKa = 3.94NYY459 pKa = 10.88LLIMRR464 pKa = 11.84DD465 pKa = 3.3LGVEE469 pKa = 3.92INLSKK474 pKa = 11.05SLISSTGVVEE484 pKa = 4.74FAKK487 pKa = 10.13RR488 pKa = 11.84WRR490 pKa = 11.84VGQTDD495 pKa = 3.66VSPASPALITRR506 pKa = 11.84LLSNMNYY513 pKa = 10.27LPVLILDD520 pKa = 4.13LVNRR524 pKa = 11.84GVKK527 pKa = 9.54TIQNSEE533 pKa = 4.18KK534 pKa = 10.53LLSLSDD540 pKa = 3.79SLHH543 pKa = 6.42KK544 pKa = 10.14IKK546 pKa = 10.79KK547 pKa = 9.98SITFSLIPYY556 pKa = 8.96YY557 pKa = 10.63DD558 pKa = 3.74SEE560 pKa = 4.35FAKK563 pKa = 10.79CLLPLKK569 pKa = 10.65GKK571 pKa = 10.21DD572 pKa = 3.05KK573 pKa = 11.26SFSQKK578 pKa = 10.14EE579 pKa = 3.86LFALYY584 pKa = 10.42VHH586 pKa = 6.66TDD588 pKa = 3.85RR589 pKa = 11.84IFNKK593 pKa = 10.27LGLLEE598 pKa = 3.8FHH600 pKa = 7.02KK601 pKa = 11.06AKK603 pKa = 10.99DD604 pKa = 3.59GDD606 pKa = 3.92QEE608 pKa = 4.12NSKK611 pKa = 10.69LSQSYY616 pKa = 10.25LYY618 pKa = 10.77NILDD622 pKa = 3.72NSKK625 pKa = 10.83LPLPAITSQIDD636 pKa = 3.57NILIAQMKK644 pKa = 8.92TITNVPTRR652 pKa = 11.84NWLNNRR658 pKa = 11.84DD659 pKa = 3.55INLLTFEE666 pKa = 4.86CWHH669 pKa = 6.87SYY671 pKa = 10.9LIDD674 pKa = 3.76TLSEE678 pKa = 3.88LSSISKK684 pKa = 9.02VVPDD688 pKa = 3.14YY689 pKa = 11.18TIIKK693 pKa = 10.41DD694 pKa = 3.67NEE696 pKa = 4.19KK697 pKa = 10.41PDD699 pKa = 3.83NLEE702 pKa = 3.45MRR704 pKa = 11.84RR705 pKa = 11.84NLLFMQEE712 pKa = 4.53LIKK715 pKa = 10.59DD716 pKa = 3.99LKK718 pKa = 10.13KK719 pKa = 9.04WNPGIFEE726 pKa = 5.45IIPEE730 pKa = 4.53SILKK734 pKa = 10.26EE735 pKa = 4.12EE736 pKa = 4.37EE737 pKa = 4.03VV738 pKa = 3.56
Molecular weight: 84.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A5YRF7|A5YRF7_9VIRU RNA-dependent RNA polymerase OS=Botrytis cinerea mitovirus 1 OX=444193 GN=RDRP PE=4 SV=3
MM1 pKa = 7.27KK2 pKa = 10.2HH3 pKa = 6.41LFRR6 pKa = 11.84IKK8 pKa = 10.14RR9 pKa = 11.84KK10 pKa = 9.69AVIKK14 pKa = 10.78NSTTRR19 pKa = 11.84LPDD22 pKa = 3.96HH23 pKa = 6.8LWLGYY28 pKa = 10.63VKK30 pKa = 10.7VLVWLYY36 pKa = 9.65DD37 pKa = 3.18IPSAHH42 pKa = 5.68RR43 pKa = 11.84TYY45 pKa = 10.8FFEE48 pKa = 4.47LLNRR52 pKa = 11.84IQSLRR57 pKa = 11.84RR58 pKa = 11.84NSGDD62 pKa = 2.81TWTVGYY68 pKa = 10.28LKK70 pKa = 10.26EE71 pKa = 4.06SHH73 pKa = 6.64RR74 pKa = 11.84LSSHH78 pKa = 5.36WLSGNPDD85 pKa = 3.21VCSGSIRR92 pKa = 11.84VSVRR96 pKa = 11.84GLPLIIPGRR105 pKa = 11.84LRR107 pKa = 11.84NVIYY111 pKa = 10.55SRR113 pKa = 11.84NNINITRR120 pKa = 11.84ALLTLLSTFRR130 pKa = 11.84VMSATPKK137 pKa = 10.49LKK139 pKa = 10.63LSTITDD145 pKa = 3.73PFNGVNEE152 pKa = 4.34TLPSLEE158 pKa = 3.91VSRR161 pKa = 11.84ALKK164 pKa = 10.74NILQGNKK171 pKa = 7.26FHH173 pKa = 7.32FKK175 pKa = 10.76KK176 pKa = 10.62EE177 pKa = 4.33DD178 pKa = 3.09IKK180 pKa = 11.45DD181 pKa = 3.69PYY183 pKa = 10.6SVNRR187 pKa = 11.84LMDD190 pKa = 3.2ITKK193 pKa = 10.1ADD195 pKa = 3.87PNGGVSLRR203 pKa = 11.84NVIIDD208 pKa = 3.97AYY210 pKa = 10.95ALMKK214 pKa = 10.0HH215 pKa = 5.83PKK217 pKa = 9.69VYY219 pKa = 10.89ASLLEE224 pKa = 4.2LSNVMSPYY232 pKa = 10.47LGSLLRR238 pKa = 11.84NQTTIIRR245 pKa = 11.84AIIDD249 pKa = 3.51YY250 pKa = 10.87DD251 pKa = 4.11PIEE254 pKa = 4.1WEE256 pKa = 4.05RR257 pKa = 11.84IKK259 pKa = 11.07PFMAPIYY266 pKa = 10.46QNIKK270 pKa = 10.69DD271 pKa = 3.84RR272 pKa = 11.84TLKK275 pKa = 10.6LVPDD279 pKa = 4.32LSCIGKK285 pKa = 10.29LSTKK289 pKa = 10.2EE290 pKa = 3.51EE291 pKa = 4.03AAGKK295 pKa = 9.74VRR297 pKa = 11.84VFAMVDD303 pKa = 2.32IWTQSILNPLHH314 pKa = 6.71KK315 pKa = 10.3KK316 pKa = 8.81IFSIIRR322 pKa = 11.84EE323 pKa = 4.43LPTDD327 pKa = 3.6GTFDD331 pKa = 3.33QLKK334 pKa = 10.04PLDD337 pKa = 4.18RR338 pKa = 11.84LHH340 pKa = 6.86EE341 pKa = 4.63LSTQDD346 pKa = 3.61RR347 pKa = 11.84FSFDD351 pKa = 3.18LSAATDD357 pKa = 3.93RR358 pKa = 11.84LPLTLQKK365 pKa = 10.86DD366 pKa = 3.44ILTLLVSPSFAEE378 pKa = 3.88AWGTALVGRR387 pKa = 11.84PYY389 pKa = 10.38KK390 pKa = 10.01WKK392 pKa = 10.49FGTTEE397 pKa = 4.12DD398 pKa = 3.49EE399 pKa = 4.12LMYY402 pKa = 10.94SVGQPMGALSSWGMLALTHH421 pKa = 5.49HH422 pKa = 7.14TIVQVAASRR431 pKa = 11.84AGYY434 pKa = 10.29KK435 pKa = 10.44DD436 pKa = 4.38LFLDD440 pKa = 3.91YY441 pKa = 11.17ALLGDD446 pKa = 5.67DD447 pKa = 3.56ICIANKK453 pKa = 10.14AVADD457 pKa = 3.94NYY459 pKa = 10.88LLIMRR464 pKa = 11.84DD465 pKa = 3.3LGVEE469 pKa = 3.92INLSKK474 pKa = 11.05SLISSTGVVEE484 pKa = 4.74FAKK487 pKa = 10.13RR488 pKa = 11.84WRR490 pKa = 11.84VGQTDD495 pKa = 3.66VSPASPALITRR506 pKa = 11.84LLSNMNYY513 pKa = 10.27LPVLILDD520 pKa = 4.13LVNRR524 pKa = 11.84GVKK527 pKa = 9.54TIQNSEE533 pKa = 4.18KK534 pKa = 10.53LLSLSDD540 pKa = 3.79SLHH543 pKa = 6.42KK544 pKa = 10.14IKK546 pKa = 10.79KK547 pKa = 9.98SITFSLIPYY556 pKa = 8.96YY557 pKa = 10.63DD558 pKa = 3.74SEE560 pKa = 4.35FAKK563 pKa = 10.79CLLPLKK569 pKa = 10.65GKK571 pKa = 10.21DD572 pKa = 3.05KK573 pKa = 11.26SFSQKK578 pKa = 10.14EE579 pKa = 3.86LFALYY584 pKa = 10.42VHH586 pKa = 6.66TDD588 pKa = 3.85RR589 pKa = 11.84IFNKK593 pKa = 10.27LGLLEE598 pKa = 3.8FHH600 pKa = 7.02KK601 pKa = 11.06AKK603 pKa = 10.99DD604 pKa = 3.59GDD606 pKa = 3.92QEE608 pKa = 4.12NSKK611 pKa = 10.69LSQSYY616 pKa = 10.25LYY618 pKa = 10.77NILDD622 pKa = 3.72NSKK625 pKa = 10.83LPLPAITSQIDD636 pKa = 3.57NILIAQMKK644 pKa = 8.92TITNVPTRR652 pKa = 11.84NWLNNRR658 pKa = 11.84DD659 pKa = 3.55INLLTFEE666 pKa = 4.86CWHH669 pKa = 6.87SYY671 pKa = 10.9LIDD674 pKa = 3.76TLSEE678 pKa = 3.88LSSISKK684 pKa = 9.02VVPDD688 pKa = 3.14YY689 pKa = 11.18TIIKK693 pKa = 10.41DD694 pKa = 3.67NEE696 pKa = 4.19KK697 pKa = 10.41PDD699 pKa = 3.83NLEE702 pKa = 3.45MRR704 pKa = 11.84RR705 pKa = 11.84NLLFMQEE712 pKa = 4.53LIKK715 pKa = 10.59DD716 pKa = 3.99LKK718 pKa = 10.13KK719 pKa = 9.04WNPGIFEE726 pKa = 5.45IIPEE730 pKa = 4.53SILKK734 pKa = 10.26EE735 pKa = 4.12EE736 pKa = 4.37EE737 pKa = 4.03VV738 pKa = 3.56
MM1 pKa = 7.27KK2 pKa = 10.2HH3 pKa = 6.41LFRR6 pKa = 11.84IKK8 pKa = 10.14RR9 pKa = 11.84KK10 pKa = 9.69AVIKK14 pKa = 10.78NSTTRR19 pKa = 11.84LPDD22 pKa = 3.96HH23 pKa = 6.8LWLGYY28 pKa = 10.63VKK30 pKa = 10.7VLVWLYY36 pKa = 9.65DD37 pKa = 3.18IPSAHH42 pKa = 5.68RR43 pKa = 11.84TYY45 pKa = 10.8FFEE48 pKa = 4.47LLNRR52 pKa = 11.84IQSLRR57 pKa = 11.84RR58 pKa = 11.84NSGDD62 pKa = 2.81TWTVGYY68 pKa = 10.28LKK70 pKa = 10.26EE71 pKa = 4.06SHH73 pKa = 6.64RR74 pKa = 11.84LSSHH78 pKa = 5.36WLSGNPDD85 pKa = 3.21VCSGSIRR92 pKa = 11.84VSVRR96 pKa = 11.84GLPLIIPGRR105 pKa = 11.84LRR107 pKa = 11.84NVIYY111 pKa = 10.55SRR113 pKa = 11.84NNINITRR120 pKa = 11.84ALLTLLSTFRR130 pKa = 11.84VMSATPKK137 pKa = 10.49LKK139 pKa = 10.63LSTITDD145 pKa = 3.73PFNGVNEE152 pKa = 4.34TLPSLEE158 pKa = 3.91VSRR161 pKa = 11.84ALKK164 pKa = 10.74NILQGNKK171 pKa = 7.26FHH173 pKa = 7.32FKK175 pKa = 10.76KK176 pKa = 10.62EE177 pKa = 4.33DD178 pKa = 3.09IKK180 pKa = 11.45DD181 pKa = 3.69PYY183 pKa = 10.6SVNRR187 pKa = 11.84LMDD190 pKa = 3.2ITKK193 pKa = 10.1ADD195 pKa = 3.87PNGGVSLRR203 pKa = 11.84NVIIDD208 pKa = 3.97AYY210 pKa = 10.95ALMKK214 pKa = 10.0HH215 pKa = 5.83PKK217 pKa = 9.69VYY219 pKa = 10.89ASLLEE224 pKa = 4.2LSNVMSPYY232 pKa = 10.47LGSLLRR238 pKa = 11.84NQTTIIRR245 pKa = 11.84AIIDD249 pKa = 3.51YY250 pKa = 10.87DD251 pKa = 4.11PIEE254 pKa = 4.1WEE256 pKa = 4.05RR257 pKa = 11.84IKK259 pKa = 11.07PFMAPIYY266 pKa = 10.46QNIKK270 pKa = 10.69DD271 pKa = 3.84RR272 pKa = 11.84TLKK275 pKa = 10.6LVPDD279 pKa = 4.32LSCIGKK285 pKa = 10.29LSTKK289 pKa = 10.2EE290 pKa = 3.51EE291 pKa = 4.03AAGKK295 pKa = 9.74VRR297 pKa = 11.84VFAMVDD303 pKa = 2.32IWTQSILNPLHH314 pKa = 6.71KK315 pKa = 10.3KK316 pKa = 8.81IFSIIRR322 pKa = 11.84EE323 pKa = 4.43LPTDD327 pKa = 3.6GTFDD331 pKa = 3.33QLKK334 pKa = 10.04PLDD337 pKa = 4.18RR338 pKa = 11.84LHH340 pKa = 6.86EE341 pKa = 4.63LSTQDD346 pKa = 3.61RR347 pKa = 11.84FSFDD351 pKa = 3.18LSAATDD357 pKa = 3.93RR358 pKa = 11.84LPLTLQKK365 pKa = 10.86DD366 pKa = 3.44ILTLLVSPSFAEE378 pKa = 3.88AWGTALVGRR387 pKa = 11.84PYY389 pKa = 10.38KK390 pKa = 10.01WKK392 pKa = 10.49FGTTEE397 pKa = 4.12DD398 pKa = 3.49EE399 pKa = 4.12LMYY402 pKa = 10.94SVGQPMGALSSWGMLALTHH421 pKa = 5.49HH422 pKa = 7.14TIVQVAASRR431 pKa = 11.84AGYY434 pKa = 10.29KK435 pKa = 10.44DD436 pKa = 4.38LFLDD440 pKa = 3.91YY441 pKa = 11.17ALLGDD446 pKa = 5.67DD447 pKa = 3.56ICIANKK453 pKa = 10.14AVADD457 pKa = 3.94NYY459 pKa = 10.88LLIMRR464 pKa = 11.84DD465 pKa = 3.3LGVEE469 pKa = 3.92INLSKK474 pKa = 11.05SLISSTGVVEE484 pKa = 4.74FAKK487 pKa = 10.13RR488 pKa = 11.84WRR490 pKa = 11.84VGQTDD495 pKa = 3.66VSPASPALITRR506 pKa = 11.84LLSNMNYY513 pKa = 10.27LPVLILDD520 pKa = 4.13LVNRR524 pKa = 11.84GVKK527 pKa = 9.54TIQNSEE533 pKa = 4.18KK534 pKa = 10.53LLSLSDD540 pKa = 3.79SLHH543 pKa = 6.42KK544 pKa = 10.14IKK546 pKa = 10.79KK547 pKa = 9.98SITFSLIPYY556 pKa = 8.96YY557 pKa = 10.63DD558 pKa = 3.74SEE560 pKa = 4.35FAKK563 pKa = 10.79CLLPLKK569 pKa = 10.65GKK571 pKa = 10.21DD572 pKa = 3.05KK573 pKa = 11.26SFSQKK578 pKa = 10.14EE579 pKa = 3.86LFALYY584 pKa = 10.42VHH586 pKa = 6.66TDD588 pKa = 3.85RR589 pKa = 11.84IFNKK593 pKa = 10.27LGLLEE598 pKa = 3.8FHH600 pKa = 7.02KK601 pKa = 11.06AKK603 pKa = 10.99DD604 pKa = 3.59GDD606 pKa = 3.92QEE608 pKa = 4.12NSKK611 pKa = 10.69LSQSYY616 pKa = 10.25LYY618 pKa = 10.77NILDD622 pKa = 3.72NSKK625 pKa = 10.83LPLPAITSQIDD636 pKa = 3.57NILIAQMKK644 pKa = 8.92TITNVPTRR652 pKa = 11.84NWLNNRR658 pKa = 11.84DD659 pKa = 3.55INLLTFEE666 pKa = 4.86CWHH669 pKa = 6.87SYY671 pKa = 10.9LIDD674 pKa = 3.76TLSEE678 pKa = 3.88LSSISKK684 pKa = 9.02VVPDD688 pKa = 3.14YY689 pKa = 11.18TIIKK693 pKa = 10.41DD694 pKa = 3.67NEE696 pKa = 4.19KK697 pKa = 10.41PDD699 pKa = 3.83NLEE702 pKa = 3.45MRR704 pKa = 11.84RR705 pKa = 11.84NLLFMQEE712 pKa = 4.53LIKK715 pKa = 10.59DD716 pKa = 3.99LKK718 pKa = 10.13KK719 pKa = 9.04WNPGIFEE726 pKa = 5.45IIPEE730 pKa = 4.53SILKK734 pKa = 10.26EE735 pKa = 4.12EE736 pKa = 4.37EE737 pKa = 4.03VV738 pKa = 3.56
Molecular weight: 84.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
738 |
738 |
738 |
738.0 |
84.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.878 ± 0.0 | 0.678 ± 0.0 |
6.098 ± 0.0 | 4.336 ± 0.0 |
3.523 ± 0.0 | 4.201 ± 0.0 |
2.033 ± 0.0 | 8.13 ± 0.0 |
7.453 ± 0.0 | 14.092 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.033 ± 0.0 | 5.42 ± 0.0 |
4.743 ± 0.0 | 2.439 ± 0.0 |
5.285 ± 0.0 | 8.537 ± 0.0 |
5.691 ± 0.0 | 5.42 ± 0.0 |
1.762 ± 0.0 | 3.252 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
