
Herpetosiphon aurantiacus (strain ATCC 23779 / DSM 785 / 114-95)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Chloroflexia; Herpetosiphonales; Herpetosiphonaceae; Herpetosiphon; Herpetosiphon aurantiacus
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
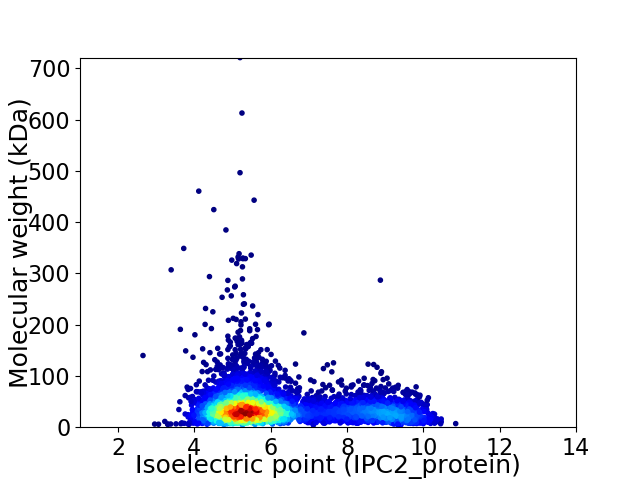
Virtual 2D-PAGE plot for 5254 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A9B7N3|A9B7N3_HERA2 Uncharacterized protein OS=Herpetosiphon aurantiacus (strain ATCC 23779 / DSM 785 / 114-95) OX=316274 GN=Haur_0355 PE=4 SV=1
MM1 pKa = 8.07RR2 pKa = 11.84LRR4 pKa = 11.84IGFIGLMIALLGLTGGLRR22 pKa = 11.84SLTSVQAVPNAPQNLAAHH40 pKa = 5.8QLNTPQGFARR50 pKa = 11.84ISEE53 pKa = 4.3QEE55 pKa = 4.33SNDD58 pKa = 3.85TPAMAQPIIGDD69 pKa = 3.66SAVIRR74 pKa = 11.84GYY76 pKa = 8.6ITANDD81 pKa = 3.89SDD83 pKa = 4.31FLAIPLTSVQRR94 pKa = 11.84VVAATMTSASATNIVDD110 pKa = 4.69SILTLYY116 pKa = 10.41QPDD119 pKa = 3.85GTTVLEE125 pKa = 4.58LDD127 pKa = 3.78NDD129 pKa = 4.09DD130 pKa = 5.22GIFGVSSSVISSAIITTTATYY151 pKa = 9.77YY152 pKa = 11.19LKK154 pKa = 9.78VTANNTTNKK163 pKa = 9.53IYY165 pKa = 10.83YY166 pKa = 7.88YY167 pKa = 10.29DD168 pKa = 3.95LYY170 pKa = 11.62VRR172 pKa = 11.84ILTSEE177 pKa = 4.71PIVEE181 pKa = 4.13QEE183 pKa = 4.17PNEE186 pKa = 4.06PAQMLPAAGIVSGIISQTGDD206 pKa = 2.66IDD208 pKa = 4.26RR209 pKa = 11.84FQLALNPGDD218 pKa = 3.86TLFTTLDD225 pKa = 3.39IDD227 pKa = 3.9PEE229 pKa = 4.25RR230 pKa = 11.84DD231 pKa = 3.62GISWNGRR238 pKa = 11.84VTIGPFGPNIITINDD253 pKa = 3.6GNAVSPNAEE262 pKa = 4.19SAQLTVKK269 pKa = 10.32EE270 pKa = 4.1AGNYY274 pKa = 9.02IISVDD279 pKa = 3.59NLSAINNSTYY289 pKa = 10.54ILQVLIIPAEE299 pKa = 4.07EE300 pKa = 3.8QANCQTYY307 pKa = 9.55MSTDD311 pKa = 3.28VNKK314 pKa = 9.95TIPPDD319 pKa = 3.68PGMITSGLNIPDD331 pKa = 3.69NLLIGDD337 pKa = 5.06LDD339 pKa = 5.24LIIQLDD345 pKa = 3.69HH346 pKa = 6.53TFIPDD351 pKa = 4.11LDD353 pKa = 4.2AQLTTPDD360 pKa = 3.98GNILGLFSDD369 pKa = 4.12IGITTSAPASMNLIIDD385 pKa = 4.44DD386 pKa = 4.07EE387 pKa = 4.47AALPSFQALSVNGLINLPEE406 pKa = 3.96AAYY409 pKa = 10.59RR410 pKa = 11.84MSWFDD415 pKa = 3.43GQRR418 pKa = 11.84TQGNWTLTLYY428 pKa = 11.03DD429 pKa = 4.72DD430 pKa = 3.91ATGDD434 pKa = 4.69GGTLLNWGLQVCAALPPADD453 pKa = 4.79CPDD456 pKa = 3.52GTVSSTIYY464 pKa = 9.88STDD467 pKa = 3.38FEE469 pKa = 6.09ANDD472 pKa = 3.93GGFTHH477 pKa = 7.13SGTTDD482 pKa = 2.84PWQYY486 pKa = 10.64GQPNSAPIVGSYY498 pKa = 10.12SGSNSWKK505 pKa = 9.31TNLTGNYY512 pKa = 7.01PALMNANLTSPAIDD526 pKa = 3.45LSGVVGPIQVQWQQRR541 pKa = 11.84YY542 pKa = 8.76QIEE545 pKa = 4.17SAAFDD550 pKa = 3.51NYY552 pKa = 10.46AAQIVGSSNQVLFQHH567 pKa = 7.02RR568 pKa = 11.84DD569 pKa = 2.97GVMRR573 pKa = 11.84EE574 pKa = 4.24SVGNPLVTVQASTGWSRR591 pKa = 11.84QLHH594 pKa = 6.76DD595 pKa = 3.2ISSFVGQSIQLQFHH609 pKa = 7.0MDD611 pKa = 3.3TDD613 pKa = 4.14SSVEE617 pKa = 3.93LAGVAIDD624 pKa = 4.29DD625 pKa = 4.13VLVTGCVVLPTATPTEE641 pKa = 4.63TPTNTPTNTPTEE653 pKa = 4.5TPTNTPTVTVTPSNTPIVTPSVTAGPTTIPVYY685 pKa = 10.72LPLVSKK691 pKa = 11.49GEE693 pKa = 4.0
MM1 pKa = 8.07RR2 pKa = 11.84LRR4 pKa = 11.84IGFIGLMIALLGLTGGLRR22 pKa = 11.84SLTSVQAVPNAPQNLAAHH40 pKa = 5.8QLNTPQGFARR50 pKa = 11.84ISEE53 pKa = 4.3QEE55 pKa = 4.33SNDD58 pKa = 3.85TPAMAQPIIGDD69 pKa = 3.66SAVIRR74 pKa = 11.84GYY76 pKa = 8.6ITANDD81 pKa = 3.89SDD83 pKa = 4.31FLAIPLTSVQRR94 pKa = 11.84VVAATMTSASATNIVDD110 pKa = 4.69SILTLYY116 pKa = 10.41QPDD119 pKa = 3.85GTTVLEE125 pKa = 4.58LDD127 pKa = 3.78NDD129 pKa = 4.09DD130 pKa = 5.22GIFGVSSSVISSAIITTTATYY151 pKa = 9.77YY152 pKa = 11.19LKK154 pKa = 9.78VTANNTTNKK163 pKa = 9.53IYY165 pKa = 10.83YY166 pKa = 7.88YY167 pKa = 10.29DD168 pKa = 3.95LYY170 pKa = 11.62VRR172 pKa = 11.84ILTSEE177 pKa = 4.71PIVEE181 pKa = 4.13QEE183 pKa = 4.17PNEE186 pKa = 4.06PAQMLPAAGIVSGIISQTGDD206 pKa = 2.66IDD208 pKa = 4.26RR209 pKa = 11.84FQLALNPGDD218 pKa = 3.86TLFTTLDD225 pKa = 3.39IDD227 pKa = 3.9PEE229 pKa = 4.25RR230 pKa = 11.84DD231 pKa = 3.62GISWNGRR238 pKa = 11.84VTIGPFGPNIITINDD253 pKa = 3.6GNAVSPNAEE262 pKa = 4.19SAQLTVKK269 pKa = 10.32EE270 pKa = 4.1AGNYY274 pKa = 9.02IISVDD279 pKa = 3.59NLSAINNSTYY289 pKa = 10.54ILQVLIIPAEE299 pKa = 4.07EE300 pKa = 3.8QANCQTYY307 pKa = 9.55MSTDD311 pKa = 3.28VNKK314 pKa = 9.95TIPPDD319 pKa = 3.68PGMITSGLNIPDD331 pKa = 3.69NLLIGDD337 pKa = 5.06LDD339 pKa = 5.24LIIQLDD345 pKa = 3.69HH346 pKa = 6.53TFIPDD351 pKa = 4.11LDD353 pKa = 4.2AQLTTPDD360 pKa = 3.98GNILGLFSDD369 pKa = 4.12IGITTSAPASMNLIIDD385 pKa = 4.44DD386 pKa = 4.07EE387 pKa = 4.47AALPSFQALSVNGLINLPEE406 pKa = 3.96AAYY409 pKa = 10.59RR410 pKa = 11.84MSWFDD415 pKa = 3.43GQRR418 pKa = 11.84TQGNWTLTLYY428 pKa = 11.03DD429 pKa = 4.72DD430 pKa = 3.91ATGDD434 pKa = 4.69GGTLLNWGLQVCAALPPADD453 pKa = 4.79CPDD456 pKa = 3.52GTVSSTIYY464 pKa = 9.88STDD467 pKa = 3.38FEE469 pKa = 6.09ANDD472 pKa = 3.93GGFTHH477 pKa = 7.13SGTTDD482 pKa = 2.84PWQYY486 pKa = 10.64GQPNSAPIVGSYY498 pKa = 10.12SGSNSWKK505 pKa = 9.31TNLTGNYY512 pKa = 7.01PALMNANLTSPAIDD526 pKa = 3.45LSGVVGPIQVQWQQRR541 pKa = 11.84YY542 pKa = 8.76QIEE545 pKa = 4.17SAAFDD550 pKa = 3.51NYY552 pKa = 10.46AAQIVGSSNQVLFQHH567 pKa = 7.02RR568 pKa = 11.84DD569 pKa = 2.97GVMRR573 pKa = 11.84EE574 pKa = 4.24SVGNPLVTVQASTGWSRR591 pKa = 11.84QLHH594 pKa = 6.76DD595 pKa = 3.2ISSFVGQSIQLQFHH609 pKa = 7.0MDD611 pKa = 3.3TDD613 pKa = 4.14SSVEE617 pKa = 3.93LAGVAIDD624 pKa = 4.29DD625 pKa = 4.13VLVTGCVVLPTATPTEE641 pKa = 4.63TPTNTPTNTPTEE653 pKa = 4.5TPTNTPTVTVTPSNTPIVTPSVTAGPTTIPVYY685 pKa = 10.72LPLVSKK691 pKa = 11.49GEE693 pKa = 4.0
Molecular weight: 73.61 kDa
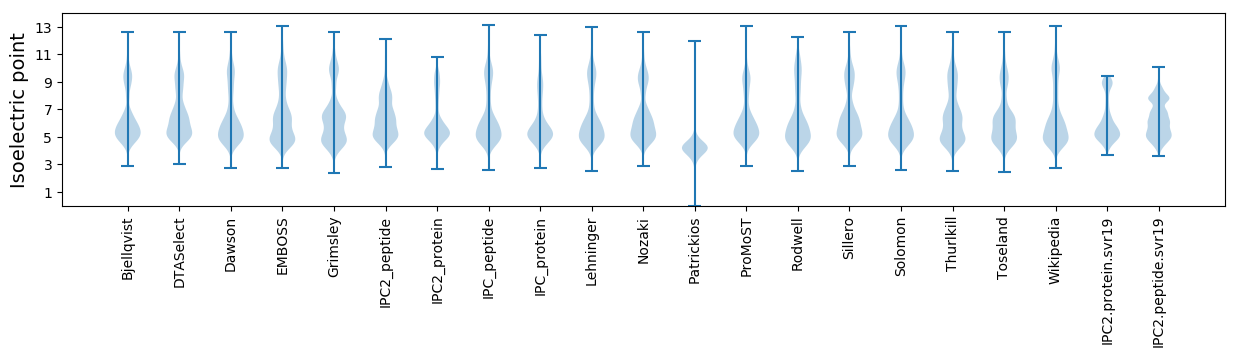
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A9AXS7|A9AXS7_HERA2 Uncharacterized protein OS=Herpetosiphon aurantiacus (strain ATCC 23779 / DSM 785 / 114-95) OX=316274 GN=Haur_2253 PE=4 SV=1
MM1 pKa = 8.05PKK3 pKa = 9.18RR4 pKa = 11.84TWQPKK9 pKa = 7.6RR10 pKa = 11.84IKK12 pKa = 9.86RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84VHH17 pKa = 6.51GFLEE21 pKa = 4.43RR22 pKa = 11.84MQTRR26 pKa = 11.84TGRR29 pKa = 11.84AVLKK33 pKa = 10.54ARR35 pKa = 11.84RR36 pKa = 11.84QRR38 pKa = 11.84GRR40 pKa = 11.84WKK42 pKa = 9.76LTVSDD47 pKa = 3.88KK48 pKa = 10.8RR49 pKa = 11.84RR50 pKa = 11.84PAGPGGHH57 pKa = 6.3RR58 pKa = 3.81
MM1 pKa = 8.05PKK3 pKa = 9.18RR4 pKa = 11.84TWQPKK9 pKa = 7.6RR10 pKa = 11.84IKK12 pKa = 9.86RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84VHH17 pKa = 6.51GFLEE21 pKa = 4.43RR22 pKa = 11.84MQTRR26 pKa = 11.84TGRR29 pKa = 11.84AVLKK33 pKa = 10.54ARR35 pKa = 11.84RR36 pKa = 11.84QRR38 pKa = 11.84GRR40 pKa = 11.84WKK42 pKa = 9.76LTVSDD47 pKa = 3.88KK48 pKa = 10.8RR49 pKa = 11.84RR50 pKa = 11.84PAGPGGHH57 pKa = 6.3RR58 pKa = 3.81
Molecular weight: 7.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1962024 |
38 |
6661 |
373.4 |
41.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.671 ± 0.045 | 0.815 ± 0.01 |
5.036 ± 0.026 | 5.167 ± 0.031 |
3.443 ± 0.021 | 7.107 ± 0.031 |
2.247 ± 0.017 | 5.993 ± 0.031 |
2.629 ± 0.025 | 11.132 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.094 ± 0.015 | 3.704 ± 0.031 |
5.165 ± 0.031 | 5.541 ± 0.034 |
5.86 ± 0.031 | 5.883 ± 0.029 |
6.088 ± 0.039 | 6.775 ± 0.032 |
1.737 ± 0.016 | 2.913 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
