
Canine oral papillomavirus (strain Y62) (COPV)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Lambdapapillomavirus; Lambdapapillomavirus 2
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
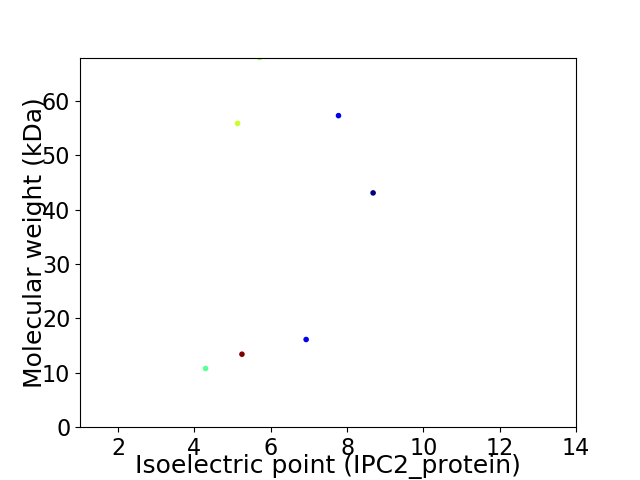
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
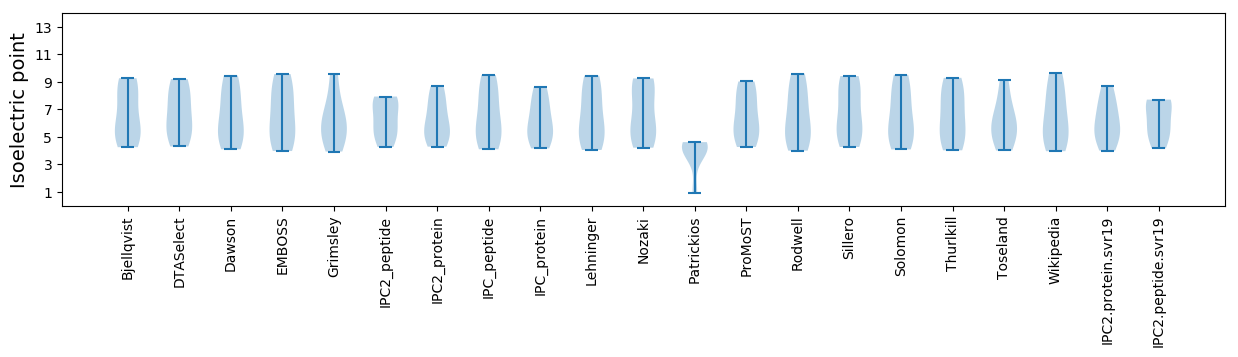
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q89808|VE6_COPV6 Protein E6 OS=Canine oral papillomavirus (strain Y62) OX=766192 GN=E6 PE=3 SV=1
MM1 pKa = 7.71IGQCATLLDD10 pKa = 3.93IVLTEE15 pKa = 4.01QPEE18 pKa = 4.57PIDD21 pKa = 3.79LQCYY25 pKa = 7.01EE26 pKa = 4.54QLPSSDD32 pKa = 3.59EE33 pKa = 4.11EE34 pKa = 4.39EE35 pKa = 4.54EE36 pKa = 4.29EE37 pKa = 4.74EE38 pKa = 5.02EE39 pKa = 4.05PTEE42 pKa = 4.14KK43 pKa = 10.45NVYY46 pKa = 9.9RR47 pKa = 11.84IEE49 pKa = 4.21AACGFCGKK57 pKa = 9.69GVRR60 pKa = 11.84FFCLSQKK67 pKa = 10.56EE68 pKa = 4.28DD69 pKa = 3.54LRR71 pKa = 11.84VLQVTLLSLSLVCTTCVQTAKK92 pKa = 10.31LDD94 pKa = 3.57HH95 pKa = 6.66GGG97 pKa = 3.36
MM1 pKa = 7.71IGQCATLLDD10 pKa = 3.93IVLTEE15 pKa = 4.01QPEE18 pKa = 4.57PIDD21 pKa = 3.79LQCYY25 pKa = 7.01EE26 pKa = 4.54QLPSSDD32 pKa = 3.59EE33 pKa = 4.11EE34 pKa = 4.39EE35 pKa = 4.54EE36 pKa = 4.29EE37 pKa = 4.74EE38 pKa = 5.02EE39 pKa = 4.05PTEE42 pKa = 4.14KK43 pKa = 10.45NVYY46 pKa = 9.9RR47 pKa = 11.84IEE49 pKa = 4.21AACGFCGKK57 pKa = 9.69GVRR60 pKa = 11.84FFCLSQKK67 pKa = 10.56EE68 pKa = 4.28DD69 pKa = 3.54LRR71 pKa = 11.84VLQVTLLSLSLVCTTCVQTAKK92 pKa = 10.31LDD94 pKa = 3.57HH95 pKa = 6.66GGG97 pKa = 3.36
Molecular weight: 10.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q89536|VE1_COPV6 Replication protein E1 OS=Canine oral papillomavirus (strain Y62) OX=766192 GN=E1 PE=3 SV=1
MM1 pKa = 7.61EE2 pKa = 5.69KK3 pKa = 10.51LSEE6 pKa = 4.4ALDD9 pKa = 3.92LLQEE13 pKa = 4.44EE14 pKa = 4.93LLSLYY19 pKa = 8.59EE20 pKa = 4.1QNSQSLADD28 pKa = 4.1QSRR31 pKa = 11.84HH32 pKa = 3.67WSLLRR37 pKa = 11.84KK38 pKa = 9.21EE39 pKa = 4.34QVLLYY44 pKa = 8.49YY45 pKa = 10.85ARR47 pKa = 11.84GKK49 pKa = 10.76GIMRR53 pKa = 11.84IGMQPVPPQSVSQAKK68 pKa = 9.62AKK70 pKa = 10.09QAIEE74 pKa = 3.58QSLYY78 pKa = 10.2IDD80 pKa = 4.01SLLHH84 pKa = 5.88SKK86 pKa = 9.78YY87 pKa = 10.94ANEE90 pKa = 3.95PWTLCDD96 pKa = 3.24TSRR99 pKa = 11.84EE100 pKa = 4.0RR101 pKa = 11.84LVAEE105 pKa = 4.0PAYY108 pKa = 9.42TFKK111 pKa = 11.02KK112 pKa = 10.11GGKK115 pKa = 9.42QIDD118 pKa = 3.7VRR120 pKa = 11.84YY121 pKa = 10.06GDD123 pKa = 3.61SEE125 pKa = 4.5EE126 pKa = 4.66NIVRR130 pKa = 11.84YY131 pKa = 8.48VLWLDD136 pKa = 4.01IYY138 pKa = 11.14YY139 pKa = 10.07QDD141 pKa = 5.89EE142 pKa = 4.17FDD144 pKa = 3.62TWEE147 pKa = 4.17KK148 pKa = 11.1AHH150 pKa = 6.75GKK152 pKa = 9.6LDD154 pKa = 3.92HH155 pKa = 6.94KK156 pKa = 10.75GLSYY160 pKa = 10.15MHH162 pKa = 6.88GTQQVYY168 pKa = 9.19YY169 pKa = 10.84VDD171 pKa = 5.16FEE173 pKa = 4.5EE174 pKa = 4.64EE175 pKa = 3.89ANKK178 pKa = 10.51YY179 pKa = 10.61SEE181 pKa = 4.08TGKK184 pKa = 10.83YY185 pKa = 9.66EE186 pKa = 4.11ILNQPTTIPTTSAAGTSGPEE206 pKa = 4.12LPGHH210 pKa = 5.73SASGSGACSLTPRR223 pKa = 11.84KK224 pKa = 10.08GPSRR228 pKa = 11.84RR229 pKa = 11.84PGRR232 pKa = 11.84RR233 pKa = 11.84SSRR236 pKa = 11.84FPRR239 pKa = 11.84RR240 pKa = 11.84SGGRR244 pKa = 11.84GRR246 pKa = 11.84LGRR249 pKa = 11.84GGSGEE254 pKa = 5.33LPPQPQPSSSWSPPSPQQVGSKK276 pKa = 9.47HH277 pKa = 4.77QLRR280 pKa = 11.84TTSSAGGRR288 pKa = 11.84LGRR291 pKa = 11.84LLQEE295 pKa = 5.1AYY297 pKa = 10.24DD298 pKa = 3.96PPVLVLAGDD307 pKa = 4.11PNSLKK312 pKa = 10.49CIRR315 pKa = 11.84YY316 pKa = 9.01RR317 pKa = 11.84LSHH320 pKa = 5.77KK321 pKa = 10.37HH322 pKa = 5.52RR323 pKa = 11.84GLYY326 pKa = 10.33LGASTTWKK334 pKa = 6.73WTSGGDD340 pKa = 3.44GASKK344 pKa = 10.34HH345 pKa = 6.38DD346 pKa = 3.92RR347 pKa = 11.84GSARR351 pKa = 11.84MLLAFLSDD359 pKa = 3.52QQRR362 pKa = 11.84EE363 pKa = 4.13DD364 pKa = 3.32FMDD367 pKa = 3.59RR368 pKa = 11.84VTFPKK373 pKa = 9.87SVRR376 pKa = 11.84VFRR379 pKa = 11.84GGLDD383 pKa = 3.34EE384 pKa = 4.83LL385 pKa = 4.75
MM1 pKa = 7.61EE2 pKa = 5.69KK3 pKa = 10.51LSEE6 pKa = 4.4ALDD9 pKa = 3.92LLQEE13 pKa = 4.44EE14 pKa = 4.93LLSLYY19 pKa = 8.59EE20 pKa = 4.1QNSQSLADD28 pKa = 4.1QSRR31 pKa = 11.84HH32 pKa = 3.67WSLLRR37 pKa = 11.84KK38 pKa = 9.21EE39 pKa = 4.34QVLLYY44 pKa = 8.49YY45 pKa = 10.85ARR47 pKa = 11.84GKK49 pKa = 10.76GIMRR53 pKa = 11.84IGMQPVPPQSVSQAKK68 pKa = 9.62AKK70 pKa = 10.09QAIEE74 pKa = 3.58QSLYY78 pKa = 10.2IDD80 pKa = 4.01SLLHH84 pKa = 5.88SKK86 pKa = 9.78YY87 pKa = 10.94ANEE90 pKa = 3.95PWTLCDD96 pKa = 3.24TSRR99 pKa = 11.84EE100 pKa = 4.0RR101 pKa = 11.84LVAEE105 pKa = 4.0PAYY108 pKa = 9.42TFKK111 pKa = 11.02KK112 pKa = 10.11GGKK115 pKa = 9.42QIDD118 pKa = 3.7VRR120 pKa = 11.84YY121 pKa = 10.06GDD123 pKa = 3.61SEE125 pKa = 4.5EE126 pKa = 4.66NIVRR130 pKa = 11.84YY131 pKa = 8.48VLWLDD136 pKa = 4.01IYY138 pKa = 11.14YY139 pKa = 10.07QDD141 pKa = 5.89EE142 pKa = 4.17FDD144 pKa = 3.62TWEE147 pKa = 4.17KK148 pKa = 11.1AHH150 pKa = 6.75GKK152 pKa = 9.6LDD154 pKa = 3.92HH155 pKa = 6.94KK156 pKa = 10.75GLSYY160 pKa = 10.15MHH162 pKa = 6.88GTQQVYY168 pKa = 9.19YY169 pKa = 10.84VDD171 pKa = 5.16FEE173 pKa = 4.5EE174 pKa = 4.64EE175 pKa = 3.89ANKK178 pKa = 10.51YY179 pKa = 10.61SEE181 pKa = 4.08TGKK184 pKa = 10.83YY185 pKa = 9.66EE186 pKa = 4.11ILNQPTTIPTTSAAGTSGPEE206 pKa = 4.12LPGHH210 pKa = 5.73SASGSGACSLTPRR223 pKa = 11.84KK224 pKa = 10.08GPSRR228 pKa = 11.84RR229 pKa = 11.84PGRR232 pKa = 11.84RR233 pKa = 11.84SSRR236 pKa = 11.84FPRR239 pKa = 11.84RR240 pKa = 11.84SGGRR244 pKa = 11.84GRR246 pKa = 11.84LGRR249 pKa = 11.84GGSGEE254 pKa = 5.33LPPQPQPSSSWSPPSPQQVGSKK276 pKa = 9.47HH277 pKa = 4.77QLRR280 pKa = 11.84TTSSAGGRR288 pKa = 11.84LGRR291 pKa = 11.84LLQEE295 pKa = 5.1AYY297 pKa = 10.24DD298 pKa = 3.96PPVLVLAGDD307 pKa = 4.11PNSLKK312 pKa = 10.49CIRR315 pKa = 11.84YY316 pKa = 9.01RR317 pKa = 11.84LSHH320 pKa = 5.77KK321 pKa = 10.37HH322 pKa = 5.52RR323 pKa = 11.84GLYY326 pKa = 10.33LGASTTWKK334 pKa = 6.73WTSGGDD340 pKa = 3.44GASKK344 pKa = 10.34HH345 pKa = 6.38DD346 pKa = 3.92RR347 pKa = 11.84GSARR351 pKa = 11.84MLLAFLSDD359 pKa = 3.52QQRR362 pKa = 11.84EE363 pKa = 4.13DD364 pKa = 3.32FMDD367 pKa = 3.59RR368 pKa = 11.84VTFPKK373 pKa = 9.87SVRR376 pKa = 11.84VFRR379 pKa = 11.84GGLDD383 pKa = 3.34EE384 pKa = 4.83LL385 pKa = 4.75
Molecular weight: 43.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2357 |
97 |
597 |
336.7 |
37.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.261 ± 0.527 | 2.249 ± 0.824 |
6.067 ± 0.411 | 6.576 ± 0.661 |
4.497 ± 0.592 | 7.0 ± 0.647 |
2.333 ± 0.161 | 4.328 ± 0.934 |
5.388 ± 0.574 | 9.504 ± 1.108 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.4 ± 0.235 | 3.521 ± 0.707 |
7.0 ± 1.232 | 4.455 ± 0.407 |
6.194 ± 0.509 | 8.401 ± 0.864 |
5.685 ± 0.373 | 5.685 ± 0.607 |
1.358 ± 0.293 | 3.097 ± 0.465 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
