
Propionibacterium phage Pirate
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Pahexavirus; Propionibacterium virus Pirate
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
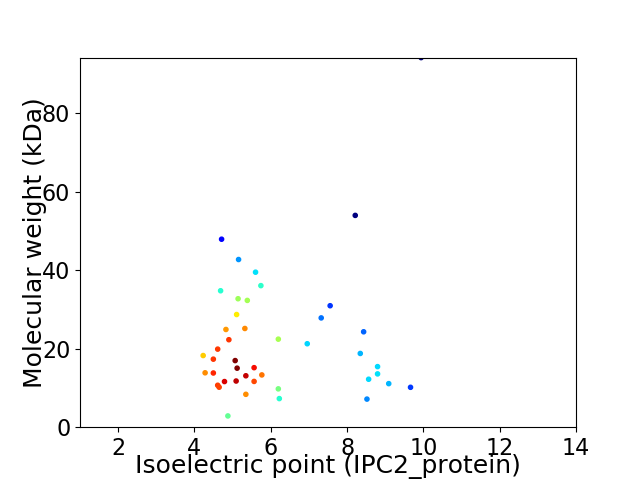
Virtual 2D-PAGE plot for 44 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
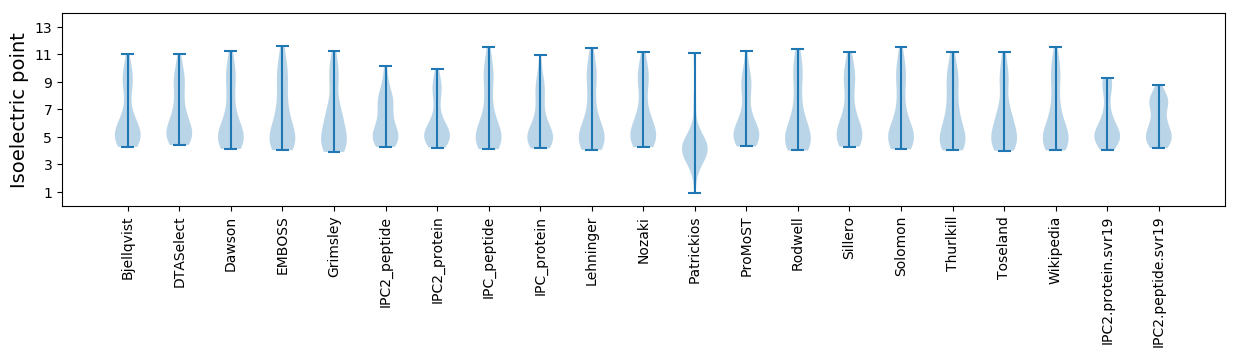
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0H4IMU4|A0A0H4IMU4_9CAUD Helix-turn-helix DNA binding domain protein OS=Propionibacterium phage Pirate OX=1655018 GN=SEA_PIRATE_27 PE=4 SV=1
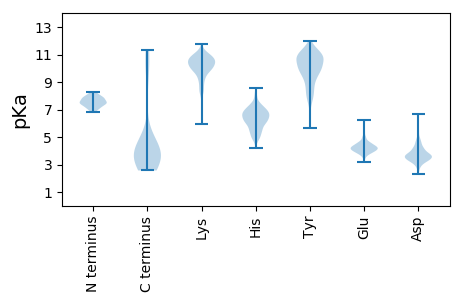
MM1 pKa = 7.48NNTHH5 pKa = 7.25NIPQAALKK13 pKa = 8.74TAVHH17 pKa = 6.82RR18 pKa = 11.84IIRR21 pKa = 11.84QQPTNMQQLEE31 pKa = 4.49NIVADD36 pKa = 3.38VEE38 pKa = 4.17NQYY41 pKa = 10.62RR42 pKa = 11.84IPISLDD48 pKa = 3.14NVNLTIKK55 pKa = 10.24EE56 pKa = 4.05VSLDD60 pKa = 3.58NLAINQDD67 pKa = 3.64TLDD70 pKa = 3.66EE71 pKa = 4.73CGEE74 pKa = 4.22ILWDD78 pKa = 4.35CDD80 pKa = 3.49SAGYY84 pKa = 6.93PTNNPQPEE92 pKa = 4.09ASQAALDD99 pKa = 3.84GLEE102 pKa = 4.02CLTYY106 pKa = 10.56KK107 pKa = 10.91ALTLITMVDD116 pKa = 3.76NILEE120 pKa = 4.85AIHH123 pKa = 6.43DD124 pKa = 3.95HH125 pKa = 6.7RR126 pKa = 11.84DD127 pKa = 3.31NYY129 pKa = 10.26PGIVKK134 pKa = 10.15QNIVDD139 pKa = 3.61QAKK142 pKa = 8.21YY143 pKa = 8.88TLAEE147 pKa = 4.37CALLHH152 pKa = 5.51QTLEE156 pKa = 5.37DD157 pKa = 3.69ILDD160 pKa = 4.26DD161 pKa = 4.06NLL163 pKa = 4.43
MM1 pKa = 7.48NNTHH5 pKa = 7.25NIPQAALKK13 pKa = 8.74TAVHH17 pKa = 6.82RR18 pKa = 11.84IIRR21 pKa = 11.84QQPTNMQQLEE31 pKa = 4.49NIVADD36 pKa = 3.38VEE38 pKa = 4.17NQYY41 pKa = 10.62RR42 pKa = 11.84IPISLDD48 pKa = 3.14NVNLTIKK55 pKa = 10.24EE56 pKa = 4.05VSLDD60 pKa = 3.58NLAINQDD67 pKa = 3.64TLDD70 pKa = 3.66EE71 pKa = 4.73CGEE74 pKa = 4.22ILWDD78 pKa = 4.35CDD80 pKa = 3.49SAGYY84 pKa = 6.93PTNNPQPEE92 pKa = 4.09ASQAALDD99 pKa = 3.84GLEE102 pKa = 4.02CLTYY106 pKa = 10.56KK107 pKa = 10.91ALTLITMVDD116 pKa = 3.76NILEE120 pKa = 4.85AIHH123 pKa = 6.43DD124 pKa = 3.95HH125 pKa = 6.7RR126 pKa = 11.84DD127 pKa = 3.31NYY129 pKa = 10.26PGIVKK134 pKa = 10.15QNIVDD139 pKa = 3.61QAKK142 pKa = 8.21YY143 pKa = 8.88TLAEE147 pKa = 4.37CALLHH152 pKa = 5.51QTLEE156 pKa = 5.37DD157 pKa = 3.69ILDD160 pKa = 4.26DD161 pKa = 4.06NLL163 pKa = 4.43
Molecular weight: 18.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0H4ISJ7|A0A0H4ISJ7_9CAUD Minor tail protein OS=Propionibacterium phage Pirate OX=1655018 GN=SEA_PIRATE_19 PE=4 SV=1
MM1 pKa = 7.85SGEE4 pKa = 3.79IASAYY9 pKa = 9.42VSLYY13 pKa = 10.73TKK15 pKa = 10.26MPGLKK20 pKa = 9.77SDD22 pKa = 3.48VGKK25 pKa = 10.2QLSGVMPAEE34 pKa = 4.46GQRR37 pKa = 11.84SGSLFAKK44 pKa = 10.08GMKK47 pKa = 9.56LALGGAAMVGAINVAKK63 pKa = 10.41KK64 pKa = 10.12GLKK67 pKa = 9.86SIYY70 pKa = 10.1DD71 pKa = 3.46VTIGGGIARR80 pKa = 11.84AMAIDD85 pKa = 3.74EE86 pKa = 4.58AQAKK90 pKa = 8.36LTGLGHH96 pKa = 6.51TSSDD100 pKa = 3.06TSSIMNSAIEE110 pKa = 4.15AVTGTSYY117 pKa = 11.95ALGDD121 pKa = 3.87AASTAAALSASGVKK135 pKa = 10.18SGGQMTDD142 pKa = 2.69VLKK145 pKa = 10.01TVADD149 pKa = 3.5VSYY152 pKa = 10.98ISGKK156 pKa = 9.96SFQDD160 pKa = 2.85TGAIFTSVMARR171 pKa = 11.84GKK173 pKa = 10.44LQGDD177 pKa = 4.11DD178 pKa = 3.62MLQLTMAGVPVLSLLARR195 pKa = 11.84QTGKK199 pKa = 9.04TSAEE203 pKa = 4.02VSQMVSKK210 pKa = 10.7GQIDD214 pKa = 4.08FATFSAAMKK223 pKa = 10.31LGMGGAAQASGKK235 pKa = 7.48TFEE238 pKa = 4.62GAMKK242 pKa = 10.13NVKK245 pKa = 9.81GALGYY250 pKa = 10.89LGATAMAPFLNGLRR264 pKa = 11.84QIFVALNPVIKK275 pKa = 10.57SVTDD279 pKa = 3.32SVKK282 pKa = 10.72PMFAAVDD289 pKa = 3.54AGIQRR294 pKa = 11.84MMPSILAWINRR305 pKa = 11.84MPGMITRR312 pKa = 11.84MNAQMRR318 pKa = 11.84AKK320 pKa = 10.19VEE322 pKa = 3.84QLKK325 pKa = 10.85SVFARR330 pKa = 11.84LHH332 pKa = 5.77LPVPKK337 pKa = 10.54VNLGAMFAGGTAVFGIVAAGVGKK360 pKa = 10.13LVAGFAPLAVSLKK373 pKa = 10.55NLLPSFGALKK383 pKa = 10.35GAAGGLGGVFRR394 pKa = 11.84ALGGPVGIVIGLFAAMFATNAQFRR418 pKa = 11.84AAVMQLVAVVGQALGQIMAAIQPLFGLVAGVVAQLAPVFGQIIGMVAGLAAQIVPLISMLVARR481 pKa = 11.84LVPVITQIIGAVTQVAAMLLPALMPVLQAVVAVIRR516 pKa = 11.84QVVGVVMQLVPVLVPVIQQILGAVMSVLPPIIGLIRR552 pKa = 11.84SLIPVIMSIMRR563 pKa = 11.84VVMQVVGVVLQVVARR578 pKa = 11.84IIPVVMPIVTAVIGFVARR596 pKa = 11.84ILGAVVSAAARR607 pKa = 11.84IIGTVTRR614 pKa = 11.84VIGWIVGHH622 pKa = 5.97FVSGVARR629 pKa = 11.84MGSVILNGWNHH640 pKa = 5.06IRR642 pKa = 11.84AFTSAFINGFKK653 pKa = 10.55SIISGGVNAVVGFFTRR669 pKa = 11.84LGLSVASHH677 pKa = 5.49VRR679 pKa = 11.84SGFNAARR686 pKa = 11.84GAVSSAMNAIRR697 pKa = 11.84SVVSSVTSAVGGFFSSMASRR717 pKa = 11.84VRR719 pKa = 11.84SGAVRR724 pKa = 11.84GFNGARR730 pKa = 11.84NAASSAMHH738 pKa = 6.4AMGSAVSSGVHH749 pKa = 5.07GVLGFFRR756 pKa = 11.84NLPGNIRR763 pKa = 11.84HH764 pKa = 6.5ALGNMGSLLVSAGRR778 pKa = 11.84DD779 pKa = 3.57VVSGLGNGIRR789 pKa = 11.84NAMSGLLDD797 pKa = 3.46TVRR800 pKa = 11.84NMGSQVANAAKK811 pKa = 10.09SVLGIHH817 pKa = 5.93SPSRR821 pKa = 11.84VFRR824 pKa = 11.84DD825 pKa = 3.43EE826 pKa = 3.91VGRR829 pKa = 11.84QVVAGLAEE837 pKa = 5.4GITGNAGLALDD848 pKa = 4.39AMSGVADD855 pKa = 4.76RR856 pKa = 11.84LPDD859 pKa = 3.5TVDD862 pKa = 2.54ARR864 pKa = 11.84FGVRR868 pKa = 11.84SSVGSFTPYY877 pKa = 10.31GRR879 pKa = 11.84YY880 pKa = 8.5QRR882 pKa = 11.84MSDD885 pKa = 2.89KK886 pKa = 11.06SVVVNVNGPTYY897 pKa = 10.88GDD899 pKa = 3.46PNEE902 pKa = 3.83FAKK905 pKa = 10.67RR906 pKa = 11.84IEE908 pKa = 4.16RR909 pKa = 11.84QQRR912 pKa = 11.84DD913 pKa = 3.73ALNALAYY920 pKa = 9.98VV921 pKa = 4.0
MM1 pKa = 7.85SGEE4 pKa = 3.79IASAYY9 pKa = 9.42VSLYY13 pKa = 10.73TKK15 pKa = 10.26MPGLKK20 pKa = 9.77SDD22 pKa = 3.48VGKK25 pKa = 10.2QLSGVMPAEE34 pKa = 4.46GQRR37 pKa = 11.84SGSLFAKK44 pKa = 10.08GMKK47 pKa = 9.56LALGGAAMVGAINVAKK63 pKa = 10.41KK64 pKa = 10.12GLKK67 pKa = 9.86SIYY70 pKa = 10.1DD71 pKa = 3.46VTIGGGIARR80 pKa = 11.84AMAIDD85 pKa = 3.74EE86 pKa = 4.58AQAKK90 pKa = 8.36LTGLGHH96 pKa = 6.51TSSDD100 pKa = 3.06TSSIMNSAIEE110 pKa = 4.15AVTGTSYY117 pKa = 11.95ALGDD121 pKa = 3.87AASTAAALSASGVKK135 pKa = 10.18SGGQMTDD142 pKa = 2.69VLKK145 pKa = 10.01TVADD149 pKa = 3.5VSYY152 pKa = 10.98ISGKK156 pKa = 9.96SFQDD160 pKa = 2.85TGAIFTSVMARR171 pKa = 11.84GKK173 pKa = 10.44LQGDD177 pKa = 4.11DD178 pKa = 3.62MLQLTMAGVPVLSLLARR195 pKa = 11.84QTGKK199 pKa = 9.04TSAEE203 pKa = 4.02VSQMVSKK210 pKa = 10.7GQIDD214 pKa = 4.08FATFSAAMKK223 pKa = 10.31LGMGGAAQASGKK235 pKa = 7.48TFEE238 pKa = 4.62GAMKK242 pKa = 10.13NVKK245 pKa = 9.81GALGYY250 pKa = 10.89LGATAMAPFLNGLRR264 pKa = 11.84QIFVALNPVIKK275 pKa = 10.57SVTDD279 pKa = 3.32SVKK282 pKa = 10.72PMFAAVDD289 pKa = 3.54AGIQRR294 pKa = 11.84MMPSILAWINRR305 pKa = 11.84MPGMITRR312 pKa = 11.84MNAQMRR318 pKa = 11.84AKK320 pKa = 10.19VEE322 pKa = 3.84QLKK325 pKa = 10.85SVFARR330 pKa = 11.84LHH332 pKa = 5.77LPVPKK337 pKa = 10.54VNLGAMFAGGTAVFGIVAAGVGKK360 pKa = 10.13LVAGFAPLAVSLKK373 pKa = 10.55NLLPSFGALKK383 pKa = 10.35GAAGGLGGVFRR394 pKa = 11.84ALGGPVGIVIGLFAAMFATNAQFRR418 pKa = 11.84AAVMQLVAVVGQALGQIMAAIQPLFGLVAGVVAQLAPVFGQIIGMVAGLAAQIVPLISMLVARR481 pKa = 11.84LVPVITQIIGAVTQVAAMLLPALMPVLQAVVAVIRR516 pKa = 11.84QVVGVVMQLVPVLVPVIQQILGAVMSVLPPIIGLIRR552 pKa = 11.84SLIPVIMSIMRR563 pKa = 11.84VVMQVVGVVLQVVARR578 pKa = 11.84IIPVVMPIVTAVIGFVARR596 pKa = 11.84ILGAVVSAAARR607 pKa = 11.84IIGTVTRR614 pKa = 11.84VIGWIVGHH622 pKa = 5.97FVSGVARR629 pKa = 11.84MGSVILNGWNHH640 pKa = 5.06IRR642 pKa = 11.84AFTSAFINGFKK653 pKa = 10.55SIISGGVNAVVGFFTRR669 pKa = 11.84LGLSVASHH677 pKa = 5.49VRR679 pKa = 11.84SGFNAARR686 pKa = 11.84GAVSSAMNAIRR697 pKa = 11.84SVVSSVTSAVGGFFSSMASRR717 pKa = 11.84VRR719 pKa = 11.84SGAVRR724 pKa = 11.84GFNGARR730 pKa = 11.84NAASSAMHH738 pKa = 6.4AMGSAVSSGVHH749 pKa = 5.07GVLGFFRR756 pKa = 11.84NLPGNIRR763 pKa = 11.84HH764 pKa = 6.5ALGNMGSLLVSAGRR778 pKa = 11.84DD779 pKa = 3.57VVSGLGNGIRR789 pKa = 11.84NAMSGLLDD797 pKa = 3.46TVRR800 pKa = 11.84NMGSQVANAAKK811 pKa = 10.09SVLGIHH817 pKa = 5.93SPSRR821 pKa = 11.84VFRR824 pKa = 11.84DD825 pKa = 3.43EE826 pKa = 3.91VGRR829 pKa = 11.84QVVAGLAEE837 pKa = 5.4GITGNAGLALDD848 pKa = 4.39AMSGVADD855 pKa = 4.76RR856 pKa = 11.84LPDD859 pKa = 3.5TVDD862 pKa = 2.54ARR864 pKa = 11.84FGVRR868 pKa = 11.84SSVGSFTPYY877 pKa = 10.31GRR879 pKa = 11.84YY880 pKa = 8.5QRR882 pKa = 11.84MSDD885 pKa = 2.89KK886 pKa = 11.06SVVVNVNGPTYY897 pKa = 10.88GDD899 pKa = 3.46PNEE902 pKa = 3.83FAKK905 pKa = 10.67RR906 pKa = 11.84IEE908 pKa = 4.16RR909 pKa = 11.84QQRR912 pKa = 11.84DD913 pKa = 3.73ALNALAYY920 pKa = 9.98VV921 pKa = 4.0
Molecular weight: 94.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8877 |
27 |
921 |
201.8 |
22.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.756 ± 0.658 | 1.183 ± 0.208 |
6.534 ± 0.52 | 4.912 ± 0.444 |
2.771 ± 0.23 | 8.471 ± 0.634 |
2.591 ± 0.423 | 4.833 ± 0.358 |
3.988 ± 0.266 | 7.728 ± 0.265 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.49 ± 0.363 | 3.898 ± 0.373 |
5.035 ± 0.353 | 4.168 ± 0.326 |
6.117 ± 0.373 | 6.646 ± 0.402 |
6.263 ± 0.601 | 7.863 ± 0.826 |
2.028 ± 0.265 | 2.726 ± 0.296 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
