
Papillibacter cinnamivorans DSM 12816
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Papillibacter; Papillibacter cinnamivorans
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
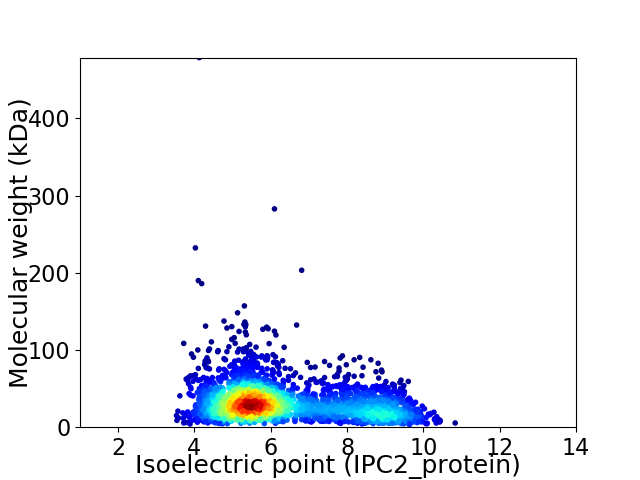
Virtual 2D-PAGE plot for 2640 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W1YI55|A0A1W1YI55_9FIRM Purine-binding chemotaxis protein CheW OS=Papillibacter cinnamivorans DSM 12816 OX=1122930 GN=SAMN02745168_0438 PE=4 SV=1
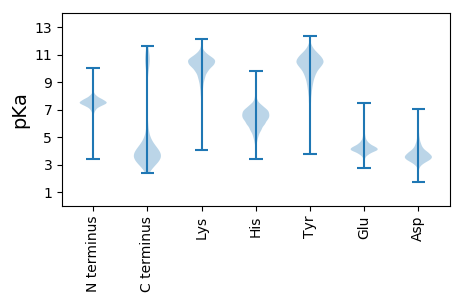
MM1 pKa = 7.67SSTFLGIQIGKK12 pKa = 9.71SGLNAAQINLNITGQNISNADD33 pKa = 3.5TQGYY37 pKa = 6.33TRR39 pKa = 11.84QSVITSSAPPSGAGYY54 pKa = 9.97VIRR57 pKa = 11.84QVTQSSNVGQGVRR70 pKa = 11.84VLSVDD75 pKa = 3.6QLRR78 pKa = 11.84SAYY81 pKa = 10.48LDD83 pKa = 3.35EE84 pKa = 4.84QYY86 pKa = 10.96RR87 pKa = 11.84SQYY90 pKa = 11.38SDD92 pKa = 3.27FCSSEE97 pKa = 3.51YY98 pKa = 9.38RR99 pKa = 11.84TQGLSYY105 pKa = 11.06LEE107 pKa = 4.71DD108 pKa = 4.3LFDD111 pKa = 4.42EE112 pKa = 5.49LDD114 pKa = 4.31DD115 pKa = 4.16NTSLTASISDD125 pKa = 3.84FFDD128 pKa = 3.69ALSDD132 pKa = 3.86FAGDD136 pKa = 3.78TTSEE140 pKa = 4.12AARR143 pKa = 11.84TTVQQTARR151 pKa = 11.84SMTDD155 pKa = 3.24NFNMIYY161 pKa = 10.89GEE163 pKa = 4.49MVDD166 pKa = 5.57LYY168 pKa = 11.01NDD170 pKa = 3.18QNTSVRR176 pKa = 11.84TVAEE180 pKa = 4.29QISSLASQIADD191 pKa = 3.92LNATICDD198 pKa = 3.88YY199 pKa = 10.99EE200 pKa = 4.92RR201 pKa = 11.84SGVTANDD208 pKa = 3.65LRR210 pKa = 11.84DD211 pKa = 3.68QRR213 pKa = 11.84NLLLDD218 pKa = 3.84KK219 pKa = 11.13LSGYY223 pKa = 11.17SDD225 pKa = 3.92FTCSEE230 pKa = 4.27DD231 pKa = 3.24AKK233 pKa = 11.35GMVNVWIAGEE243 pKa = 3.94ALVDD247 pKa = 3.89GKK249 pKa = 8.79TAGSISITSAADD261 pKa = 4.06EE262 pKa = 4.81IDD264 pKa = 3.75TLCQQLSILNGGVLSAGIITPDD286 pKa = 3.87QEE288 pKa = 4.26TQRR291 pKa = 11.84DD292 pKa = 4.93GICSALQQISGKK304 pKa = 10.02ISCNVNADD312 pKa = 3.21GTASVSIDD320 pKa = 3.53YY321 pKa = 10.68VSASMTQVTDD331 pKa = 3.57SLVDD335 pKa = 3.86GSTCGSVSEE344 pKa = 4.37EE345 pKa = 3.66AVKK348 pKa = 10.94AFDD351 pKa = 3.87GADD354 pKa = 3.36MEE356 pKa = 5.12SVLKK360 pKa = 11.0LGEE363 pKa = 4.44TYY365 pKa = 11.09LNTDD369 pKa = 3.53TVSGGEE375 pKa = 4.07LFAHH379 pKa = 6.95LSLRR383 pKa = 11.84GGDD386 pKa = 3.54TSGDD390 pKa = 3.28AGVPYY395 pKa = 10.76YY396 pKa = 10.0IGRR399 pKa = 11.84LDD401 pKa = 3.73DD402 pKa = 3.83LARR405 pKa = 11.84TVAEE409 pKa = 4.28TVNEE413 pKa = 4.13CMNKK417 pKa = 9.88GYY419 pKa = 9.1TYY421 pKa = 10.34PDD423 pKa = 3.55EE424 pKa = 4.81EE425 pKa = 4.74NGFSSVTGTDD435 pKa = 2.52VDD437 pKa = 4.33LFADD441 pKa = 4.96FGNQYY446 pKa = 11.74ALVTAGNFSVSDD458 pKa = 3.9SVLSSVWNIAGSDD471 pKa = 3.91SEE473 pKa = 4.96IDD475 pKa = 3.67LSTDD479 pKa = 2.97STQTSNNKK487 pKa = 8.95VALLLADD494 pKa = 5.62LINNTDD500 pKa = 3.72YY501 pKa = 11.96GDD503 pKa = 3.87MLDD506 pKa = 4.52GLISHH511 pKa = 7.69LGQTVSGSQSLLDD524 pKa = 3.58TRR526 pKa = 11.84EE527 pKa = 4.01SLVEE531 pKa = 3.78STEE534 pKa = 3.86NQRR537 pKa = 11.84QSISGVSVDD546 pKa = 3.97EE547 pKa = 4.3EE548 pKa = 4.33AVNLIMYY555 pKa = 7.22QQTYY559 pKa = 8.23NACSRR564 pKa = 11.84VITTMDD570 pKa = 3.01QMLDD574 pKa = 3.22KK575 pKa = 10.66LINGTGTVGLL585 pKa = 4.32
MM1 pKa = 7.67SSTFLGIQIGKK12 pKa = 9.71SGLNAAQINLNITGQNISNADD33 pKa = 3.5TQGYY37 pKa = 6.33TRR39 pKa = 11.84QSVITSSAPPSGAGYY54 pKa = 9.97VIRR57 pKa = 11.84QVTQSSNVGQGVRR70 pKa = 11.84VLSVDD75 pKa = 3.6QLRR78 pKa = 11.84SAYY81 pKa = 10.48LDD83 pKa = 3.35EE84 pKa = 4.84QYY86 pKa = 10.96RR87 pKa = 11.84SQYY90 pKa = 11.38SDD92 pKa = 3.27FCSSEE97 pKa = 3.51YY98 pKa = 9.38RR99 pKa = 11.84TQGLSYY105 pKa = 11.06LEE107 pKa = 4.71DD108 pKa = 4.3LFDD111 pKa = 4.42EE112 pKa = 5.49LDD114 pKa = 4.31DD115 pKa = 4.16NTSLTASISDD125 pKa = 3.84FFDD128 pKa = 3.69ALSDD132 pKa = 3.86FAGDD136 pKa = 3.78TTSEE140 pKa = 4.12AARR143 pKa = 11.84TTVQQTARR151 pKa = 11.84SMTDD155 pKa = 3.24NFNMIYY161 pKa = 10.89GEE163 pKa = 4.49MVDD166 pKa = 5.57LYY168 pKa = 11.01NDD170 pKa = 3.18QNTSVRR176 pKa = 11.84TVAEE180 pKa = 4.29QISSLASQIADD191 pKa = 3.92LNATICDD198 pKa = 3.88YY199 pKa = 10.99EE200 pKa = 4.92RR201 pKa = 11.84SGVTANDD208 pKa = 3.65LRR210 pKa = 11.84DD211 pKa = 3.68QRR213 pKa = 11.84NLLLDD218 pKa = 3.84KK219 pKa = 11.13LSGYY223 pKa = 11.17SDD225 pKa = 3.92FTCSEE230 pKa = 4.27DD231 pKa = 3.24AKK233 pKa = 11.35GMVNVWIAGEE243 pKa = 3.94ALVDD247 pKa = 3.89GKK249 pKa = 8.79TAGSISITSAADD261 pKa = 4.06EE262 pKa = 4.81IDD264 pKa = 3.75TLCQQLSILNGGVLSAGIITPDD286 pKa = 3.87QEE288 pKa = 4.26TQRR291 pKa = 11.84DD292 pKa = 4.93GICSALQQISGKK304 pKa = 10.02ISCNVNADD312 pKa = 3.21GTASVSIDD320 pKa = 3.53YY321 pKa = 10.68VSASMTQVTDD331 pKa = 3.57SLVDD335 pKa = 3.86GSTCGSVSEE344 pKa = 4.37EE345 pKa = 3.66AVKK348 pKa = 10.94AFDD351 pKa = 3.87GADD354 pKa = 3.36MEE356 pKa = 5.12SVLKK360 pKa = 11.0LGEE363 pKa = 4.44TYY365 pKa = 11.09LNTDD369 pKa = 3.53TVSGGEE375 pKa = 4.07LFAHH379 pKa = 6.95LSLRR383 pKa = 11.84GGDD386 pKa = 3.54TSGDD390 pKa = 3.28AGVPYY395 pKa = 10.76YY396 pKa = 10.0IGRR399 pKa = 11.84LDD401 pKa = 3.73DD402 pKa = 3.83LARR405 pKa = 11.84TVAEE409 pKa = 4.28TVNEE413 pKa = 4.13CMNKK417 pKa = 9.88GYY419 pKa = 9.1TYY421 pKa = 10.34PDD423 pKa = 3.55EE424 pKa = 4.81EE425 pKa = 4.74NGFSSVTGTDD435 pKa = 2.52VDD437 pKa = 4.33LFADD441 pKa = 4.96FGNQYY446 pKa = 11.74ALVTAGNFSVSDD458 pKa = 3.9SVLSSVWNIAGSDD471 pKa = 3.91SEE473 pKa = 4.96IDD475 pKa = 3.67LSTDD479 pKa = 2.97STQTSNNKK487 pKa = 8.95VALLLADD494 pKa = 5.62LINNTDD500 pKa = 3.72YY501 pKa = 11.96GDD503 pKa = 3.87MLDD506 pKa = 4.52GLISHH511 pKa = 7.69LGQTVSGSQSLLDD524 pKa = 3.58TRR526 pKa = 11.84EE527 pKa = 4.01SLVEE531 pKa = 3.78STEE534 pKa = 3.86NQRR537 pKa = 11.84QSISGVSVDD546 pKa = 3.97EE547 pKa = 4.3EE548 pKa = 4.33AVNLIMYY555 pKa = 7.22QQTYY559 pKa = 8.23NACSRR564 pKa = 11.84VITTMDD570 pKa = 3.01QMLDD574 pKa = 3.22KK575 pKa = 10.66LINGTGTVGLL585 pKa = 4.32
Molecular weight: 62.36 kDa
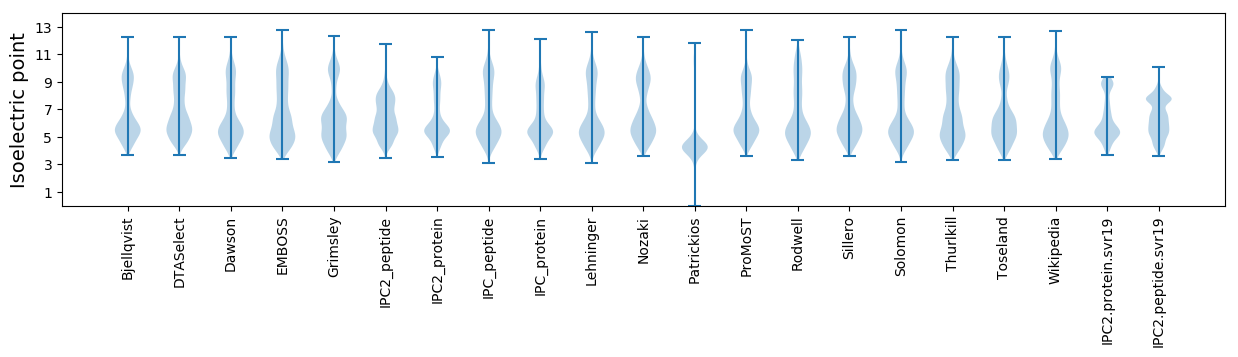
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W1Z0F6|A0A1W1Z0F6_9FIRM Stage II sporulation protein R OS=Papillibacter cinnamivorans DSM 12816 OX=1122930 GN=SAMN02745168_0836 PE=4 SV=1
MM1 pKa = 7.74WLYY4 pKa = 11.28KK5 pKa = 10.34LGRR8 pKa = 11.84SFRR11 pKa = 11.84YY12 pKa = 9.17AGRR15 pKa = 11.84GFAHH19 pKa = 6.11VLRR22 pKa = 11.84RR23 pKa = 11.84EE24 pKa = 3.98RR25 pKa = 11.84NLRR28 pKa = 11.84IHH30 pKa = 6.36LTAVFYY36 pKa = 10.16VSWAGILGRR45 pKa = 11.84LGGAEE50 pKa = 3.97WALLWICFAMVIGAEE65 pKa = 4.15LFNTAIEE72 pKa = 4.31TLGNAVTRR80 pKa = 11.84EE81 pKa = 4.02KK82 pKa = 11.33NEE84 pKa = 4.17TVGKK88 pKa = 10.29AKK90 pKa = 10.46DD91 pKa = 3.59VAAGAVLVCAVGSVGAAAGIFGCTGAWKK119 pKa = 8.72TVLRR123 pKa = 11.84SLGSSTPLSVIAIATVPAAILFIRR147 pKa = 11.84GRR149 pKa = 11.84GTWKK153 pKa = 10.6
MM1 pKa = 7.74WLYY4 pKa = 11.28KK5 pKa = 10.34LGRR8 pKa = 11.84SFRR11 pKa = 11.84YY12 pKa = 9.17AGRR15 pKa = 11.84GFAHH19 pKa = 6.11VLRR22 pKa = 11.84RR23 pKa = 11.84EE24 pKa = 3.98RR25 pKa = 11.84NLRR28 pKa = 11.84IHH30 pKa = 6.36LTAVFYY36 pKa = 10.16VSWAGILGRR45 pKa = 11.84LGGAEE50 pKa = 3.97WALLWICFAMVIGAEE65 pKa = 4.15LFNTAIEE72 pKa = 4.31TLGNAVTRR80 pKa = 11.84EE81 pKa = 4.02KK82 pKa = 11.33NEE84 pKa = 4.17TVGKK88 pKa = 10.29AKK90 pKa = 10.46DD91 pKa = 3.59VAAGAVLVCAVGSVGAAAGIFGCTGAWKK119 pKa = 8.72TVLRR123 pKa = 11.84SLGSSTPLSVIAIATVPAAILFIRR147 pKa = 11.84GRR149 pKa = 11.84GTWKK153 pKa = 10.6
Molecular weight: 16.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
814393 |
29 |
4547 |
308.5 |
33.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.244 ± 0.049 | 1.421 ± 0.02 |
5.24 ± 0.031 | 6.855 ± 0.066 |
4.005 ± 0.038 | 8.224 ± 0.051 |
1.552 ± 0.018 | 6.286 ± 0.047 |
5.067 ± 0.045 | 9.893 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.68 ± 0.025 | 3.43 ± 0.029 |
4.119 ± 0.032 | 2.72 ± 0.03 |
5.735 ± 0.063 | 6.45 ± 0.054 |
5.548 ± 0.067 | 7.126 ± 0.047 |
0.89 ± 0.018 | 3.514 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
