
Taiyuan leafhopper virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Jingchuvirales; Aliusviridae; Ollusvirus; Ollusvirus taiyuanense
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
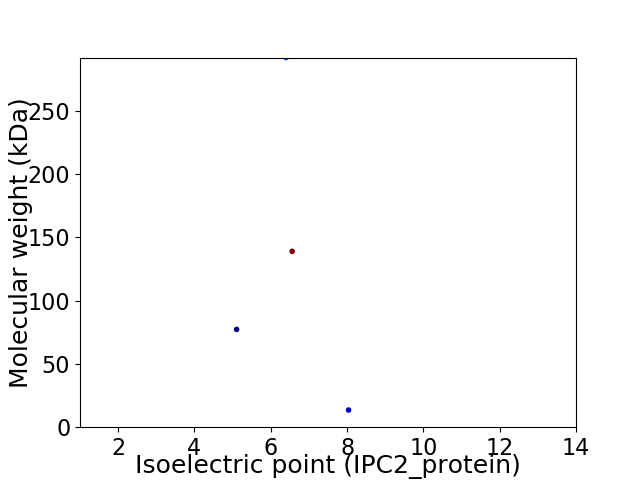
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
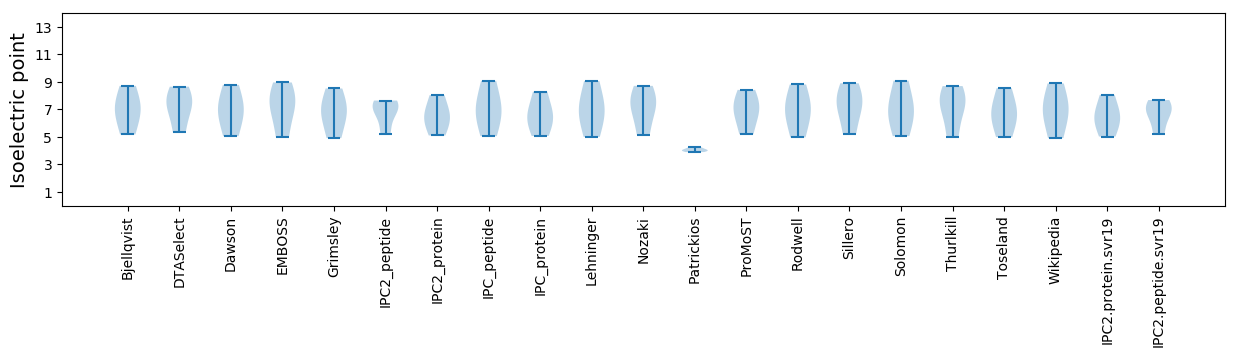
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A455LU23|A0A455LU23_9VIRU Large structural protein OS=Taiyuan leafhopper virus OX=2482723 PE=4 SV=1
MM1 pKa = 7.61ARR3 pKa = 11.84QYY5 pKa = 11.58LNAARR10 pKa = 11.84GRR12 pKa = 11.84DD13 pKa = 3.37AGVSQLIAQLNQAVSEE29 pKa = 4.16RR30 pKa = 11.84SEE32 pKa = 4.36DD33 pKa = 3.49LTPITLVAGGDD44 pKa = 3.85NIRR47 pKa = 11.84DD48 pKa = 3.59FAVAAVSGQPLDD60 pKa = 3.22IRR62 pKa = 11.84ARR64 pKa = 11.84VPLAYY69 pKa = 9.72KK70 pKa = 10.32LATYY74 pKa = 10.07FNSRR78 pKa = 11.84LSNPTPDD85 pKa = 3.5FLQARR90 pKa = 11.84LWGDD94 pKa = 3.14LHH96 pKa = 6.96AVWFVGEE103 pKa = 4.28GTNDD107 pKa = 3.26QRR109 pKa = 11.84ISANARR115 pKa = 11.84LDD117 pKa = 3.83SILIANPIEE126 pKa = 4.48GPMMLQAFQKK136 pKa = 10.79VYY138 pKa = 9.62DD139 pKa = 3.91TVRR142 pKa = 11.84AAEE145 pKa = 4.24GAGNIPWDD153 pKa = 3.72EE154 pKa = 4.06ASAVVPLAPDD164 pKa = 3.56AQDD167 pKa = 4.18LPNEE171 pKa = 4.32FPLQKK176 pKa = 10.46YY177 pKa = 8.78RR178 pKa = 11.84FFEE181 pKa = 4.49ASAHH185 pKa = 6.06AFDD188 pKa = 4.02NTALVQVTAATQDD201 pKa = 3.29ALASLRR207 pKa = 11.84EE208 pKa = 4.09INEE211 pKa = 3.89NAVNVPPHH219 pKa = 6.19NFVLNEE225 pKa = 3.85AWPQNWARR233 pKa = 11.84LSRR236 pKa = 11.84ASRR239 pKa = 11.84LACFEE244 pKa = 4.12MLYY247 pKa = 8.74HH248 pKa = 5.78TNKK251 pKa = 9.82VEE253 pKa = 3.68WDD255 pKa = 3.71SYY257 pKa = 11.2LVASVAGVVFSICKK271 pKa = 9.41SSGMTTAYY279 pKa = 10.49LQTRR283 pKa = 11.84VDD285 pKa = 4.45RR286 pKa = 11.84LVKK289 pKa = 10.25DD290 pKa = 3.78YY291 pKa = 11.39PEE293 pKa = 6.44LEE295 pKa = 4.32LQDD298 pKa = 3.96KK299 pKa = 11.22LMMEE303 pKa = 4.85FFMTFGKK310 pKa = 10.64LYY312 pKa = 10.69VPQNLDD318 pKa = 3.06RR319 pKa = 11.84TVIANYY325 pKa = 9.33LQAVYY330 pKa = 10.52GCLEE334 pKa = 4.34EE335 pKa = 5.06GPSRR339 pKa = 11.84SIAWVIEE346 pKa = 3.65QARR349 pKa = 11.84GHH351 pKa = 6.44NYY353 pKa = 7.61THH355 pKa = 7.26ALAFAEE361 pKa = 4.67SCSKK365 pKa = 9.83TKK367 pKa = 10.45YY368 pKa = 10.67LPIAYY373 pKa = 8.76LANIRR378 pKa = 11.84EE379 pKa = 4.27DD380 pKa = 3.14QWIGFIKK387 pKa = 10.25VCYY390 pKa = 9.23HH391 pKa = 5.58VCRR394 pKa = 11.84DD395 pKa = 3.35PWGSLTGPAIPANNYY410 pKa = 9.47PDD412 pKa = 3.47IANIGAIMSKK422 pKa = 10.58KK423 pKa = 9.63LDD425 pKa = 3.4KK426 pKa = 10.69AARR429 pKa = 11.84GYY431 pKa = 10.53AGKK434 pKa = 10.34FILGGEE440 pKa = 4.17HH441 pKa = 6.57TEE443 pKa = 4.23PFCKK447 pKa = 10.08AVADD451 pKa = 4.21QIIEE455 pKa = 4.03SHH457 pKa = 6.09AASLEE462 pKa = 4.29DD463 pKa = 4.02EE464 pKa = 4.79LSAANIIAALFPGIKK479 pKa = 8.75MAVVGDD485 pKa = 3.29NAYY488 pKa = 10.83YY489 pKa = 10.31YY490 pKa = 10.41DD491 pKa = 4.11EE492 pKa = 5.22SGLPLEE498 pKa = 4.7LTAEE502 pKa = 4.27NARR505 pKa = 11.84PVGGWEE511 pKa = 4.15EE512 pKa = 4.23EE513 pKa = 4.38EE514 pKa = 4.47EE515 pKa = 4.11PAPAAGAAARR525 pKa = 11.84PAAGFPGNVNRR536 pKa = 11.84EE537 pKa = 3.42VAYY540 pKa = 10.5RR541 pKa = 11.84GEE543 pKa = 4.1QPLPQIAEE551 pKa = 4.07DD552 pKa = 3.68ALAEE556 pKa = 5.04SRR558 pKa = 11.84ATWANKK564 pKa = 9.43AKK566 pKa = 10.21NLPRR570 pKa = 11.84NAVKK574 pKa = 8.09MTMKK578 pKa = 10.65DD579 pKa = 3.2QVLLLEE585 pKa = 4.97EE586 pKa = 4.47YY587 pKa = 11.13VDD589 pKa = 3.96TDD591 pKa = 3.21RR592 pKa = 11.84HH593 pKa = 5.34LSAIKK598 pKa = 9.11TCCNLLLYY606 pKa = 10.45ASKK609 pKa = 10.49QSKK612 pKa = 10.11LEE614 pKa = 4.16PYY616 pKa = 10.24GGAGDD621 pKa = 4.1NIRR624 pKa = 11.84VKK626 pKa = 10.57NRR628 pKa = 11.84YY629 pKa = 7.62VASPAGLDD637 pKa = 3.16NALNVWGLDD646 pKa = 3.66GNDD649 pKa = 4.48IIPDD653 pKa = 3.58IPEE656 pKa = 4.35DD657 pKa = 4.08APNVPYY663 pKa = 10.88NEE665 pKa = 3.72NVMGNVALYY674 pKa = 9.84ARR676 pKa = 11.84PIRR679 pKa = 11.84PIVQNRR685 pKa = 11.84RR686 pKa = 11.84GNAGLGGMLNWQPPQQNLL704 pKa = 3.1
MM1 pKa = 7.61ARR3 pKa = 11.84QYY5 pKa = 11.58LNAARR10 pKa = 11.84GRR12 pKa = 11.84DD13 pKa = 3.37AGVSQLIAQLNQAVSEE29 pKa = 4.16RR30 pKa = 11.84SEE32 pKa = 4.36DD33 pKa = 3.49LTPITLVAGGDD44 pKa = 3.85NIRR47 pKa = 11.84DD48 pKa = 3.59FAVAAVSGQPLDD60 pKa = 3.22IRR62 pKa = 11.84ARR64 pKa = 11.84VPLAYY69 pKa = 9.72KK70 pKa = 10.32LATYY74 pKa = 10.07FNSRR78 pKa = 11.84LSNPTPDD85 pKa = 3.5FLQARR90 pKa = 11.84LWGDD94 pKa = 3.14LHH96 pKa = 6.96AVWFVGEE103 pKa = 4.28GTNDD107 pKa = 3.26QRR109 pKa = 11.84ISANARR115 pKa = 11.84LDD117 pKa = 3.83SILIANPIEE126 pKa = 4.48GPMMLQAFQKK136 pKa = 10.79VYY138 pKa = 9.62DD139 pKa = 3.91TVRR142 pKa = 11.84AAEE145 pKa = 4.24GAGNIPWDD153 pKa = 3.72EE154 pKa = 4.06ASAVVPLAPDD164 pKa = 3.56AQDD167 pKa = 4.18LPNEE171 pKa = 4.32FPLQKK176 pKa = 10.46YY177 pKa = 8.78RR178 pKa = 11.84FFEE181 pKa = 4.49ASAHH185 pKa = 6.06AFDD188 pKa = 4.02NTALVQVTAATQDD201 pKa = 3.29ALASLRR207 pKa = 11.84EE208 pKa = 4.09INEE211 pKa = 3.89NAVNVPPHH219 pKa = 6.19NFVLNEE225 pKa = 3.85AWPQNWARR233 pKa = 11.84LSRR236 pKa = 11.84ASRR239 pKa = 11.84LACFEE244 pKa = 4.12MLYY247 pKa = 8.74HH248 pKa = 5.78TNKK251 pKa = 9.82VEE253 pKa = 3.68WDD255 pKa = 3.71SYY257 pKa = 11.2LVASVAGVVFSICKK271 pKa = 9.41SSGMTTAYY279 pKa = 10.49LQTRR283 pKa = 11.84VDD285 pKa = 4.45RR286 pKa = 11.84LVKK289 pKa = 10.25DD290 pKa = 3.78YY291 pKa = 11.39PEE293 pKa = 6.44LEE295 pKa = 4.32LQDD298 pKa = 3.96KK299 pKa = 11.22LMMEE303 pKa = 4.85FFMTFGKK310 pKa = 10.64LYY312 pKa = 10.69VPQNLDD318 pKa = 3.06RR319 pKa = 11.84TVIANYY325 pKa = 9.33LQAVYY330 pKa = 10.52GCLEE334 pKa = 4.34EE335 pKa = 5.06GPSRR339 pKa = 11.84SIAWVIEE346 pKa = 3.65QARR349 pKa = 11.84GHH351 pKa = 6.44NYY353 pKa = 7.61THH355 pKa = 7.26ALAFAEE361 pKa = 4.67SCSKK365 pKa = 9.83TKK367 pKa = 10.45YY368 pKa = 10.67LPIAYY373 pKa = 8.76LANIRR378 pKa = 11.84EE379 pKa = 4.27DD380 pKa = 3.14QWIGFIKK387 pKa = 10.25VCYY390 pKa = 9.23HH391 pKa = 5.58VCRR394 pKa = 11.84DD395 pKa = 3.35PWGSLTGPAIPANNYY410 pKa = 9.47PDD412 pKa = 3.47IANIGAIMSKK422 pKa = 10.58KK423 pKa = 9.63LDD425 pKa = 3.4KK426 pKa = 10.69AARR429 pKa = 11.84GYY431 pKa = 10.53AGKK434 pKa = 10.34FILGGEE440 pKa = 4.17HH441 pKa = 6.57TEE443 pKa = 4.23PFCKK447 pKa = 10.08AVADD451 pKa = 4.21QIIEE455 pKa = 4.03SHH457 pKa = 6.09AASLEE462 pKa = 4.29DD463 pKa = 4.02EE464 pKa = 4.79LSAANIIAALFPGIKK479 pKa = 8.75MAVVGDD485 pKa = 3.29NAYY488 pKa = 10.83YY489 pKa = 10.31YY490 pKa = 10.41DD491 pKa = 4.11EE492 pKa = 5.22SGLPLEE498 pKa = 4.7LTAEE502 pKa = 4.27NARR505 pKa = 11.84PVGGWEE511 pKa = 4.15EE512 pKa = 4.23EE513 pKa = 4.38EE514 pKa = 4.47EE515 pKa = 4.11PAPAAGAAARR525 pKa = 11.84PAAGFPGNVNRR536 pKa = 11.84EE537 pKa = 3.42VAYY540 pKa = 10.5RR541 pKa = 11.84GEE543 pKa = 4.1QPLPQIAEE551 pKa = 4.07DD552 pKa = 3.68ALAEE556 pKa = 5.04SRR558 pKa = 11.84ATWANKK564 pKa = 9.43AKK566 pKa = 10.21NLPRR570 pKa = 11.84NAVKK574 pKa = 8.09MTMKK578 pKa = 10.65DD579 pKa = 3.2QVLLLEE585 pKa = 4.97EE586 pKa = 4.47YY587 pKa = 11.13VDD589 pKa = 3.96TDD591 pKa = 3.21RR592 pKa = 11.84HH593 pKa = 5.34LSAIKK598 pKa = 9.11TCCNLLLYY606 pKa = 10.45ASKK609 pKa = 10.49QSKK612 pKa = 10.11LEE614 pKa = 4.16PYY616 pKa = 10.24GGAGDD621 pKa = 4.1NIRR624 pKa = 11.84VKK626 pKa = 10.57NRR628 pKa = 11.84YY629 pKa = 7.62VASPAGLDD637 pKa = 3.16NALNVWGLDD646 pKa = 3.66GNDD649 pKa = 4.48IIPDD653 pKa = 3.58IPEE656 pKa = 4.35DD657 pKa = 4.08APNVPYY663 pKa = 10.88NEE665 pKa = 3.72NVMGNVALYY674 pKa = 9.84ARR676 pKa = 11.84PIRR679 pKa = 11.84PIVQNRR685 pKa = 11.84RR686 pKa = 11.84GNAGLGGMLNWQPPQQNLL704 pKa = 3.1
Molecular weight: 77.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A455LK58|A0A455LK58_9VIRU Nucleocapsid protein OS=Taiyuan leafhopper virus OX=2482723 PE=4 SV=1
MM1 pKa = 7.72EE2 pKa = 5.72KK3 pKa = 9.4FTSYY7 pKa = 10.98SEE9 pKa = 4.05QFVYY13 pKa = 10.62LCQIYY18 pKa = 10.03HH19 pKa = 7.28LYY21 pKa = 9.91LIAMSEE27 pKa = 4.39SKK29 pKa = 10.72QITSVTQARR38 pKa = 11.84SAALSSGSVVPTNGNQQISRR58 pKa = 11.84SEE60 pKa = 3.81AMMALASDD68 pKa = 3.81ISVIKK73 pKa = 10.36SDD75 pKa = 3.38VMVIKK80 pKa = 10.33EE81 pKa = 4.55ALTKK85 pKa = 8.67IQEE88 pKa = 4.39MIGGAMKK95 pKa = 8.4TQPHH99 pKa = 7.03APPPSYY105 pKa = 11.09DD106 pKa = 3.09SGIYY110 pKa = 9.63PKK112 pKa = 10.28LHH114 pKa = 6.14RR115 pKa = 11.84PAYY118 pKa = 10.32LGGPRR123 pKa = 11.84II124 pKa = 3.99
MM1 pKa = 7.72EE2 pKa = 5.72KK3 pKa = 9.4FTSYY7 pKa = 10.98SEE9 pKa = 4.05QFVYY13 pKa = 10.62LCQIYY18 pKa = 10.03HH19 pKa = 7.28LYY21 pKa = 9.91LIAMSEE27 pKa = 4.39SKK29 pKa = 10.72QITSVTQARR38 pKa = 11.84SAALSSGSVVPTNGNQQISRR58 pKa = 11.84SEE60 pKa = 3.81AMMALASDD68 pKa = 3.81ISVIKK73 pKa = 10.36SDD75 pKa = 3.38VMVIKK80 pKa = 10.33EE81 pKa = 4.55ALTKK85 pKa = 8.67IQEE88 pKa = 4.39MIGGAMKK95 pKa = 8.4TQPHH99 pKa = 7.03APPPSYY105 pKa = 11.09DD106 pKa = 3.09SGIYY110 pKa = 9.63PKK112 pKa = 10.28LHH114 pKa = 6.14RR115 pKa = 11.84PAYY118 pKa = 10.32LGGPRR123 pKa = 11.84II124 pKa = 3.99
Molecular weight: 13.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4604 |
124 |
2546 |
1151.0 |
130.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.777 ± 1.901 | 1.781 ± 0.199 |
5.061 ± 0.288 | 5.387 ± 0.39 |
3.844 ± 0.591 | 5.039 ± 0.399 |
3.215 ± 0.512 | 6.581 ± 0.427 |
3.975 ± 0.374 | 10.904 ± 0.754 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.824 ± 0.35 | 5.169 ± 0.519 |
4.93 ± 0.419 | 4.67 ± 0.223 |
5.864 ± 0.707 | 6.407 ± 1.314 |
6.103 ± 0.89 | 6.581 ± 0.301 |
1.586 ± 0.176 | 3.301 ± 0.362 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
