
Lake Sarah-associated circular virus-47
Taxonomy: Viruses; unclassified viruses
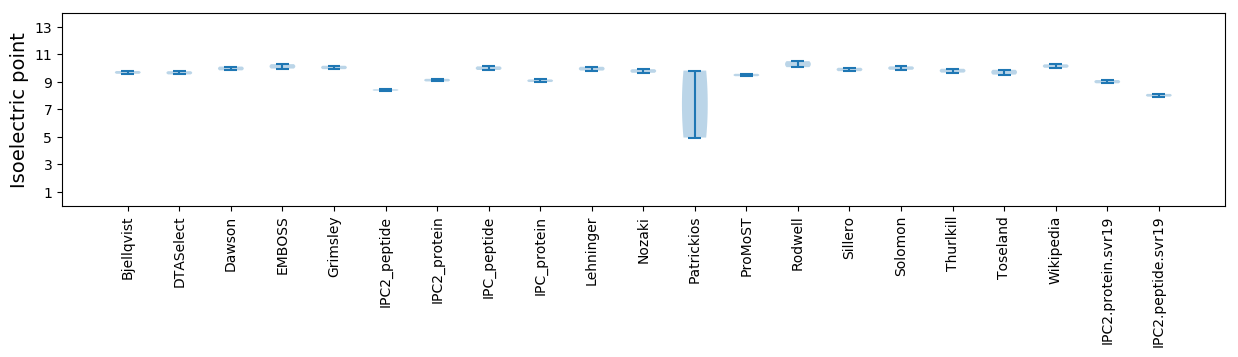
Average proteome isoelectric point is 9.02
Get precalculated fractions of proteins
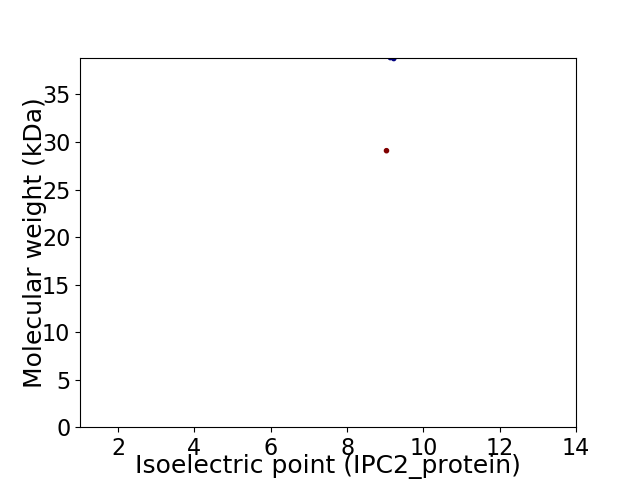
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126GAF5|A0A126GAF5_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-47 OX=1685776 PE=4 SV=1
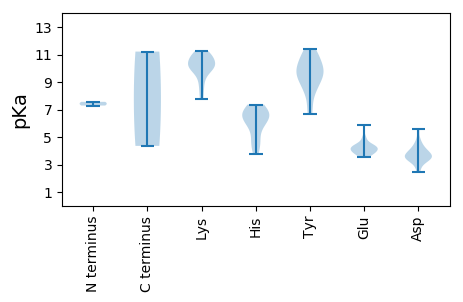
MM1 pKa = 7.29RR2 pKa = 11.84QSKK5 pKa = 9.83GRR7 pKa = 11.84AQGGVKK13 pKa = 10.09NRR15 pKa = 11.84TSGATQSRR23 pKa = 11.84KK24 pKa = 9.87GKK26 pKa = 10.51AIDD29 pKa = 3.54AVVSLRR35 pKa = 11.84QASRR39 pKa = 11.84RR40 pKa = 11.84QNQVIDD46 pKa = 3.5ATSAYY51 pKa = 8.61MLYY54 pKa = 9.54SAPMRR59 pKa = 11.84RR60 pKa = 11.84TDD62 pKa = 4.6NKK64 pKa = 10.38FFDD67 pKa = 3.49PWVANGLKK75 pKa = 10.08PIIYY79 pKa = 8.21STEE82 pKa = 4.01GAVSNSAGGYY92 pKa = 8.42VLNTAATPSACVINQIPQGTTTNSRR117 pKa = 11.84LGRR120 pKa = 11.84KK121 pKa = 8.94ARR123 pKa = 11.84ITGVYY128 pKa = 9.31IKK130 pKa = 10.76GVVQSPGAVAAAAKK144 pKa = 7.87CTLALVHH151 pKa = 6.32QLPPNNATQMPAFTDD166 pKa = 2.45IWTAQHH172 pKa = 6.84CSAQRR177 pKa = 11.84NVDD180 pKa = 3.53NNDD183 pKa = 2.8KK184 pKa = 10.77FKK186 pKa = 10.86VIRR189 pKa = 11.84AITVDD194 pKa = 3.54CMGNAAAPATGKK206 pKa = 10.27EE207 pKa = 4.37LINLDD212 pKa = 4.01EE213 pKa = 4.9MIDD216 pKa = 3.73LKK218 pKa = 11.04GKK220 pKa = 10.06EE221 pKa = 4.93IITEE225 pKa = 3.94WTAADD230 pKa = 3.58TTGVYY235 pKa = 11.25GNMEE239 pKa = 4.36RR240 pKa = 11.84GALHH244 pKa = 7.36LYY246 pKa = 10.61ALANVPFATGYY257 pKa = 9.8VFQGTVRR264 pKa = 11.84VYY266 pKa = 10.87FEE268 pKa = 5.88DD269 pKa = 4.18YY270 pKa = 11.22
MM1 pKa = 7.29RR2 pKa = 11.84QSKK5 pKa = 9.83GRR7 pKa = 11.84AQGGVKK13 pKa = 10.09NRR15 pKa = 11.84TSGATQSRR23 pKa = 11.84KK24 pKa = 9.87GKK26 pKa = 10.51AIDD29 pKa = 3.54AVVSLRR35 pKa = 11.84QASRR39 pKa = 11.84RR40 pKa = 11.84QNQVIDD46 pKa = 3.5ATSAYY51 pKa = 8.61MLYY54 pKa = 9.54SAPMRR59 pKa = 11.84RR60 pKa = 11.84TDD62 pKa = 4.6NKK64 pKa = 10.38FFDD67 pKa = 3.49PWVANGLKK75 pKa = 10.08PIIYY79 pKa = 8.21STEE82 pKa = 4.01GAVSNSAGGYY92 pKa = 8.42VLNTAATPSACVINQIPQGTTTNSRR117 pKa = 11.84LGRR120 pKa = 11.84KK121 pKa = 8.94ARR123 pKa = 11.84ITGVYY128 pKa = 9.31IKK130 pKa = 10.76GVVQSPGAVAAAAKK144 pKa = 7.87CTLALVHH151 pKa = 6.32QLPPNNATQMPAFTDD166 pKa = 2.45IWTAQHH172 pKa = 6.84CSAQRR177 pKa = 11.84NVDD180 pKa = 3.53NNDD183 pKa = 2.8KK184 pKa = 10.77FKK186 pKa = 10.86VIRR189 pKa = 11.84AITVDD194 pKa = 3.54CMGNAAAPATGKK206 pKa = 10.27EE207 pKa = 4.37LINLDD212 pKa = 4.01EE213 pKa = 4.9MIDD216 pKa = 3.73LKK218 pKa = 11.04GKK220 pKa = 10.06EE221 pKa = 4.93IITEE225 pKa = 3.94WTAADD230 pKa = 3.58TTGVYY235 pKa = 11.25GNMEE239 pKa = 4.36RR240 pKa = 11.84GALHH244 pKa = 7.36LYY246 pKa = 10.61ALANVPFATGYY257 pKa = 9.8VFQGTVRR264 pKa = 11.84VYY266 pKa = 10.87FEE268 pKa = 5.88DD269 pKa = 4.18YY270 pKa = 11.22
Molecular weight: 29.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126GAF5|A0A126GAF5_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-47 OX=1685776 PE=4 SV=1
MM1 pKa = 7.56IFGAFLTGRR10 pKa = 11.84GPQSTHH16 pKa = 3.75WCFTVHH22 pKa = 6.81RR23 pKa = 11.84PQRR26 pKa = 11.84RR27 pKa = 11.84PAVLPEE33 pKa = 3.85SCQYY37 pKa = 11.4LVFQIEE43 pKa = 4.0KK44 pKa = 7.77TAQGKK49 pKa = 8.45RR50 pKa = 11.84HH51 pKa = 4.55VQAYY55 pKa = 7.15VQFPVRR61 pKa = 11.84TRR63 pKa = 11.84GTTVSSLAKK72 pKa = 9.85RR73 pKa = 11.84IFGGEE78 pKa = 3.85PSHH81 pKa = 6.81NEE83 pKa = 3.53PAKK86 pKa = 10.98GSDD89 pKa = 3.4QEE91 pKa = 4.41NEE93 pKa = 4.38VYY95 pKa = 8.86CTKK98 pKa = 10.31EE99 pKa = 3.69ASRR102 pKa = 11.84LEE104 pKa = 4.32GPWRR108 pKa = 11.84YY109 pKa = 10.11GEE111 pKa = 4.26RR112 pKa = 11.84VPHH115 pKa = 6.44AGKK118 pKa = 10.52KK119 pKa = 9.9GGRR122 pKa = 11.84SDD124 pKa = 4.68LLKK127 pKa = 11.18LKK129 pKa = 11.03DD130 pKa = 4.03NVDD133 pKa = 3.34KK134 pKa = 11.24GATEE138 pKa = 3.92LALFEE143 pKa = 4.18QQFGDD148 pKa = 3.59MVRR151 pKa = 11.84HH152 pKa = 5.82HH153 pKa = 6.83RR154 pKa = 11.84AIGHH158 pKa = 4.42YY159 pKa = 9.65RR160 pKa = 11.84YY161 pKa = 10.41LKK163 pKa = 10.72FNVEE167 pKa = 4.31RR168 pKa = 11.84HH169 pKa = 4.98WEE171 pKa = 4.26TIVILIVGPTNTQKK185 pKa = 9.48TFLSEE190 pKa = 3.64VLARR194 pKa = 11.84SGYY197 pKa = 9.4FGKK200 pKa = 8.53TVWRR204 pKa = 11.84APEE207 pKa = 4.17SKK209 pKa = 10.6GSGMYY214 pKa = 9.57FDD216 pKa = 5.57GYY218 pKa = 10.93NGHH221 pKa = 6.63EE222 pKa = 4.26CVLWQDD228 pKa = 3.96FDD230 pKa = 4.09GGSCKK235 pKa = 10.49FGTFKK240 pKa = 11.16NLIQGTPTNVPVHH253 pKa = 6.12GAASIEE259 pKa = 3.89WAPRR263 pKa = 11.84VMILTSNYY271 pKa = 9.34LPKK274 pKa = 10.49YY275 pKa = 6.67WWKK278 pKa = 10.05SHH280 pKa = 5.88QGANDD285 pKa = 3.56LAAIYY290 pKa = 10.22KK291 pKa = 9.34RR292 pKa = 11.84VHH294 pKa = 6.46VIFKK298 pKa = 10.2RR299 pKa = 11.84LKK301 pKa = 9.18PVEE304 pKa = 3.97NMPRR308 pKa = 11.84VAPIFHH314 pKa = 7.3PASLYY319 pKa = 10.08PPRR322 pKa = 11.84PDD324 pKa = 3.27PPLAVKK330 pKa = 9.86PLAISKK336 pKa = 10.11SKK338 pKa = 10.99KK339 pKa = 10.17LFNN342 pKa = 4.36
MM1 pKa = 7.56IFGAFLTGRR10 pKa = 11.84GPQSTHH16 pKa = 3.75WCFTVHH22 pKa = 6.81RR23 pKa = 11.84PQRR26 pKa = 11.84RR27 pKa = 11.84PAVLPEE33 pKa = 3.85SCQYY37 pKa = 11.4LVFQIEE43 pKa = 4.0KK44 pKa = 7.77TAQGKK49 pKa = 8.45RR50 pKa = 11.84HH51 pKa = 4.55VQAYY55 pKa = 7.15VQFPVRR61 pKa = 11.84TRR63 pKa = 11.84GTTVSSLAKK72 pKa = 9.85RR73 pKa = 11.84IFGGEE78 pKa = 3.85PSHH81 pKa = 6.81NEE83 pKa = 3.53PAKK86 pKa = 10.98GSDD89 pKa = 3.4QEE91 pKa = 4.41NEE93 pKa = 4.38VYY95 pKa = 8.86CTKK98 pKa = 10.31EE99 pKa = 3.69ASRR102 pKa = 11.84LEE104 pKa = 4.32GPWRR108 pKa = 11.84YY109 pKa = 10.11GEE111 pKa = 4.26RR112 pKa = 11.84VPHH115 pKa = 6.44AGKK118 pKa = 10.52KK119 pKa = 9.9GGRR122 pKa = 11.84SDD124 pKa = 4.68LLKK127 pKa = 11.18LKK129 pKa = 11.03DD130 pKa = 4.03NVDD133 pKa = 3.34KK134 pKa = 11.24GATEE138 pKa = 3.92LALFEE143 pKa = 4.18QQFGDD148 pKa = 3.59MVRR151 pKa = 11.84HH152 pKa = 5.82HH153 pKa = 6.83RR154 pKa = 11.84AIGHH158 pKa = 4.42YY159 pKa = 9.65RR160 pKa = 11.84YY161 pKa = 10.41LKK163 pKa = 10.72FNVEE167 pKa = 4.31RR168 pKa = 11.84HH169 pKa = 4.98WEE171 pKa = 4.26TIVILIVGPTNTQKK185 pKa = 9.48TFLSEE190 pKa = 3.64VLARR194 pKa = 11.84SGYY197 pKa = 9.4FGKK200 pKa = 8.53TVWRR204 pKa = 11.84APEE207 pKa = 4.17SKK209 pKa = 10.6GSGMYY214 pKa = 9.57FDD216 pKa = 5.57GYY218 pKa = 10.93NGHH221 pKa = 6.63EE222 pKa = 4.26CVLWQDD228 pKa = 3.96FDD230 pKa = 4.09GGSCKK235 pKa = 10.49FGTFKK240 pKa = 11.16NLIQGTPTNVPVHH253 pKa = 6.12GAASIEE259 pKa = 3.89WAPRR263 pKa = 11.84VMILTSNYY271 pKa = 9.34LPKK274 pKa = 10.49YY275 pKa = 6.67WWKK278 pKa = 10.05SHH280 pKa = 5.88QGANDD285 pKa = 3.56LAAIYY290 pKa = 10.22KK291 pKa = 9.34RR292 pKa = 11.84VHH294 pKa = 6.46VIFKK298 pKa = 10.2RR299 pKa = 11.84LKK301 pKa = 9.18PVEE304 pKa = 3.97NMPRR308 pKa = 11.84VAPIFHH314 pKa = 7.3PASLYY319 pKa = 10.08PPRR322 pKa = 11.84PDD324 pKa = 3.27PPLAVKK330 pKa = 9.86PLAISKK336 pKa = 10.11SKK338 pKa = 10.99KK339 pKa = 10.17LFNN342 pKa = 4.36
Molecular weight: 38.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
612 |
270 |
342 |
306.0 |
33.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.641 ± 1.97 | 1.471 ± 0.006 |
3.595 ± 0.453 | 4.085 ± 0.796 |
4.085 ± 0.796 | 8.333 ± 0.099 |
2.778 ± 0.889 | 4.739 ± 0.436 |
6.373 ± 0.633 | 6.209 ± 0.546 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.961 ± 0.337 | 4.739 ± 0.831 |
5.882 ± 0.965 | 4.575 ± 0.325 |
6.373 ± 0.238 | 5.392 ± 0.11 |
6.699 ± 0.971 | 7.516 ± 0.139 |
1.797 ± 0.366 | 3.758 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
