
Haloarcula hispanica pleomorphic virus 2
Taxonomy: Viruses; Monodnaviria; Trapavirae; Saleviricota; Huolimaviricetes; Haloruvirales; Pleolipoviridae; Alphapleolipovirus; Alphapleolipovirus HHPV2
Average proteome isoelectric point is 4.95
Get precalculated fractions of proteins
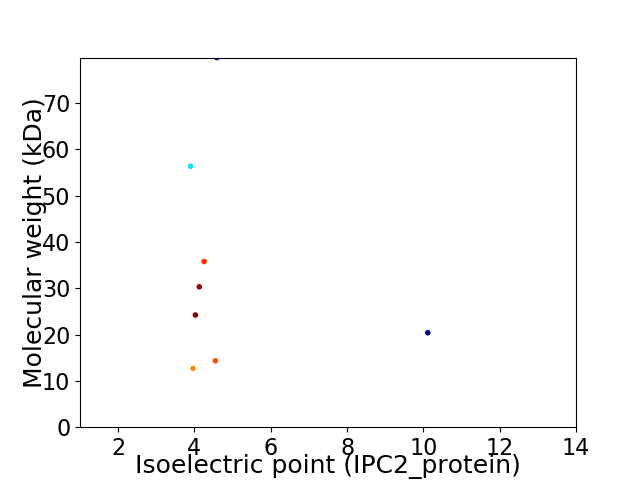
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W0FES0|W0FES0_9VIRU Gp7 OS=Haloarcula hispanica pleomorphic virus 2 OX=1442594 PE=4 SV=1
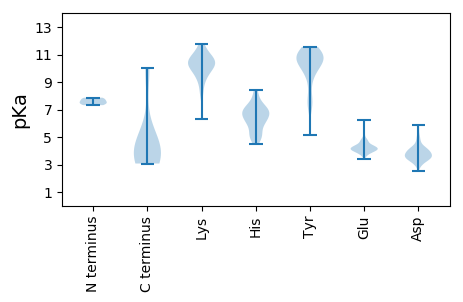
MM1 pKa = 7.86SDD3 pKa = 3.45TEE5 pKa = 4.33ASSSDD10 pKa = 2.97SSPRR14 pKa = 11.84MGRR17 pKa = 11.84RR18 pKa = 11.84TFLQTVGVTAGAAAGVKK35 pKa = 8.87YY36 pKa = 9.4TDD38 pKa = 3.65GTALDD43 pKa = 4.21PVGEE47 pKa = 4.28ADD49 pKa = 4.19AVVPLALAGVALAGAAGVGYY69 pKa = 10.75VGGSYY74 pKa = 11.01LEE76 pKa = 4.38DD77 pKa = 4.13KK78 pKa = 11.09YY79 pKa = 11.49LGDD82 pKa = 3.65SRR84 pKa = 11.84DD85 pKa = 3.5YY86 pKa = 11.32SGYY89 pKa = 9.14TGSDD93 pKa = 3.11ALKK96 pKa = 10.69SAIQEE101 pKa = 4.35GATSMKK107 pKa = 10.46VADD110 pKa = 4.6EE111 pKa = 4.25KK112 pKa = 11.8VMASIQNNIANSQNAGLAKK131 pKa = 10.21GKK133 pKa = 10.2AAIIKK138 pKa = 9.24EE139 pKa = 4.12MNAGNGEE146 pKa = 4.35SAATTAMQSSINEE159 pKa = 3.77YY160 pKa = 10.08FAAIQQNILTHH171 pKa = 6.3YY172 pKa = 7.31NTQLEE177 pKa = 4.55QVKK180 pKa = 9.65HH181 pKa = 6.2HH182 pKa = 6.61VDD184 pKa = 3.32QVANHH189 pKa = 6.35SNLTVGTVFASEE201 pKa = 4.6NGDD204 pKa = 3.63DD205 pKa = 4.3SPSNMTVNQRR215 pKa = 11.84SLSLVDD221 pKa = 3.65GTDD224 pKa = 3.02ATEE227 pKa = 4.47KK228 pKa = 10.86YY229 pKa = 10.83LIFNTSQDD237 pKa = 2.84GSTYY241 pKa = 10.58EE242 pKa = 4.1SGISLTTATGAADD255 pKa = 3.77FEE257 pKa = 4.75VDD259 pKa = 3.99SFSLSSGAYY268 pKa = 9.42ADD270 pKa = 4.3YY271 pKa = 11.02SDD273 pKa = 4.39EE274 pKa = 4.05YY275 pKa = 11.2QYY277 pKa = 11.34FQTVPMGNAFDD288 pKa = 4.54AVVTEE293 pKa = 4.11RR294 pKa = 11.84DD295 pKa = 3.34KK296 pKa = 11.67VLNDD300 pKa = 3.66LSTFVSDD307 pKa = 4.69VYY309 pKa = 11.0SQYY312 pKa = 11.33SAGDD316 pKa = 3.41IPTEE320 pKa = 4.79DD321 pKa = 3.11IVDD324 pKa = 5.43PITAATEE331 pKa = 3.59LRR333 pKa = 11.84TNYY336 pKa = 10.73DD337 pKa = 3.19GMQGASAHH345 pKa = 6.03AAMLGIPTNAEE356 pKa = 3.38QSVYY360 pKa = 7.89MTLEE364 pKa = 3.91SDD366 pKa = 5.1GVDD369 pKa = 2.87VWADD373 pKa = 2.99VFTNKK378 pKa = 10.32KK379 pKa = 7.97ITDD382 pKa = 3.58GSGNEE387 pKa = 3.78VGFQKK392 pKa = 10.12GTTYY396 pKa = 11.31DD397 pKa = 3.43PTTWDD402 pKa = 3.72DD403 pKa = 3.62PLYY406 pKa = 10.51IAFEE410 pKa = 4.19YY411 pKa = 11.09SEE413 pKa = 4.32TTDD416 pKa = 3.64ADD418 pKa = 4.11GNVLNSTQQDD428 pKa = 4.02TYY430 pKa = 11.55DD431 pKa = 3.99GEE433 pKa = 4.77TTTTEE438 pKa = 3.86YY439 pKa = 11.32SDD441 pKa = 4.05FVAVEE446 pKa = 4.02QPFTITEE453 pKa = 3.86ITVDD457 pKa = 3.88GEE459 pKa = 4.38SQQSFKK465 pKa = 8.68TTSRR469 pKa = 11.84NTQTSDD475 pKa = 2.98VSKK478 pKa = 10.57LQEE481 pKa = 4.07EE482 pKa = 4.77LDD484 pKa = 4.21QIRR487 pKa = 11.84QTQLEE492 pKa = 4.4LQEE495 pKa = 4.57EE496 pKa = 4.63SQSGGGGFDD505 pKa = 4.36LSGLSLGGIPGEE517 pKa = 4.13GVVLIAAGAAAYY529 pKa = 10.43LFGKK533 pKa = 10.04
MM1 pKa = 7.86SDD3 pKa = 3.45TEE5 pKa = 4.33ASSSDD10 pKa = 2.97SSPRR14 pKa = 11.84MGRR17 pKa = 11.84RR18 pKa = 11.84TFLQTVGVTAGAAAGVKK35 pKa = 8.87YY36 pKa = 9.4TDD38 pKa = 3.65GTALDD43 pKa = 4.21PVGEE47 pKa = 4.28ADD49 pKa = 4.19AVVPLALAGVALAGAAGVGYY69 pKa = 10.75VGGSYY74 pKa = 11.01LEE76 pKa = 4.38DD77 pKa = 4.13KK78 pKa = 11.09YY79 pKa = 11.49LGDD82 pKa = 3.65SRR84 pKa = 11.84DD85 pKa = 3.5YY86 pKa = 11.32SGYY89 pKa = 9.14TGSDD93 pKa = 3.11ALKK96 pKa = 10.69SAIQEE101 pKa = 4.35GATSMKK107 pKa = 10.46VADD110 pKa = 4.6EE111 pKa = 4.25KK112 pKa = 11.8VMASIQNNIANSQNAGLAKK131 pKa = 10.21GKK133 pKa = 10.2AAIIKK138 pKa = 9.24EE139 pKa = 4.12MNAGNGEE146 pKa = 4.35SAATTAMQSSINEE159 pKa = 3.77YY160 pKa = 10.08FAAIQQNILTHH171 pKa = 6.3YY172 pKa = 7.31NTQLEE177 pKa = 4.55QVKK180 pKa = 9.65HH181 pKa = 6.2HH182 pKa = 6.61VDD184 pKa = 3.32QVANHH189 pKa = 6.35SNLTVGTVFASEE201 pKa = 4.6NGDD204 pKa = 3.63DD205 pKa = 4.3SPSNMTVNQRR215 pKa = 11.84SLSLVDD221 pKa = 3.65GTDD224 pKa = 3.02ATEE227 pKa = 4.47KK228 pKa = 10.86YY229 pKa = 10.83LIFNTSQDD237 pKa = 2.84GSTYY241 pKa = 10.58EE242 pKa = 4.1SGISLTTATGAADD255 pKa = 3.77FEE257 pKa = 4.75VDD259 pKa = 3.99SFSLSSGAYY268 pKa = 9.42ADD270 pKa = 4.3YY271 pKa = 11.02SDD273 pKa = 4.39EE274 pKa = 4.05YY275 pKa = 11.2QYY277 pKa = 11.34FQTVPMGNAFDD288 pKa = 4.54AVVTEE293 pKa = 4.11RR294 pKa = 11.84DD295 pKa = 3.34KK296 pKa = 11.67VLNDD300 pKa = 3.66LSTFVSDD307 pKa = 4.69VYY309 pKa = 11.0SQYY312 pKa = 11.33SAGDD316 pKa = 3.41IPTEE320 pKa = 4.79DD321 pKa = 3.11IVDD324 pKa = 5.43PITAATEE331 pKa = 3.59LRR333 pKa = 11.84TNYY336 pKa = 10.73DD337 pKa = 3.19GMQGASAHH345 pKa = 6.03AAMLGIPTNAEE356 pKa = 3.38QSVYY360 pKa = 7.89MTLEE364 pKa = 3.91SDD366 pKa = 5.1GVDD369 pKa = 2.87VWADD373 pKa = 2.99VFTNKK378 pKa = 10.32KK379 pKa = 7.97ITDD382 pKa = 3.58GSGNEE387 pKa = 3.78VGFQKK392 pKa = 10.12GTTYY396 pKa = 11.31DD397 pKa = 3.43PTTWDD402 pKa = 3.72DD403 pKa = 3.62PLYY406 pKa = 10.51IAFEE410 pKa = 4.19YY411 pKa = 11.09SEE413 pKa = 4.32TTDD416 pKa = 3.64ADD418 pKa = 4.11GNVLNSTQQDD428 pKa = 4.02TYY430 pKa = 11.55DD431 pKa = 3.99GEE433 pKa = 4.77TTTTEE438 pKa = 3.86YY439 pKa = 11.32SDD441 pKa = 4.05FVAVEE446 pKa = 4.02QPFTITEE453 pKa = 3.86ITVDD457 pKa = 3.88GEE459 pKa = 4.38SQQSFKK465 pKa = 8.68TTSRR469 pKa = 11.84NTQTSDD475 pKa = 2.98VSKK478 pKa = 10.57LQEE481 pKa = 4.07EE482 pKa = 4.77LDD484 pKa = 4.21QIRR487 pKa = 11.84QTQLEE492 pKa = 4.4LQEE495 pKa = 4.57EE496 pKa = 4.63SQSGGGGFDD505 pKa = 4.36LSGLSLGGIPGEE517 pKa = 4.13GVVLIAAGAAAYY529 pKa = 10.43LFGKK533 pKa = 10.04
Molecular weight: 56.32 kDa
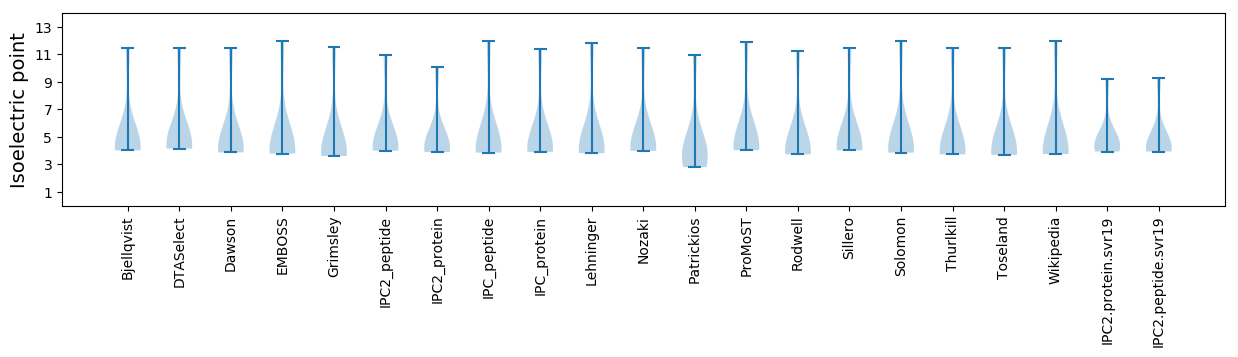
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W0FJS3|W0FJS3_9VIRU Gp8 OS=Haloarcula hispanica pleomorphic virus 2 OX=1442594 PE=4 SV=1
MM1 pKa = 7.48LRR3 pKa = 11.84TSAYY7 pKa = 10.12RR8 pKa = 11.84IYY10 pKa = 10.42TRR12 pKa = 11.84PTTAITLIPTLFISIRR28 pKa = 11.84IKK30 pKa = 10.8CLTNFRR36 pKa = 11.84NAVIAFMLEE45 pKa = 3.66RR46 pKa = 11.84RR47 pKa = 11.84KK48 pKa = 10.38LIPVGAGRR56 pKa = 11.84TLVTHH61 pKa = 7.11RR62 pKa = 11.84GFPRR66 pKa = 11.84QRR68 pKa = 11.84PVLPHH73 pKa = 7.5RR74 pKa = 11.84ILQWQLTLTKK84 pKa = 9.82LLPHH88 pKa = 6.45ATINVPDD95 pKa = 3.74NRR97 pKa = 11.84ITANLALAQEE107 pKa = 4.67TPYY110 pKa = 10.93SRR112 pKa = 11.84SQARR116 pKa = 11.84SEE118 pKa = 4.05TGIPLAHH125 pKa = 5.84RR126 pKa = 11.84TVVKK130 pKa = 9.41PHH132 pKa = 4.98QEE134 pKa = 3.74RR135 pKa = 11.84YY136 pKa = 7.04STHH139 pKa = 5.49IHH141 pKa = 5.37TQAQDD146 pKa = 3.27TSRR149 pKa = 11.84GPTGGLLSGVLWVGGFVGGNPHH171 pKa = 4.5QQEE174 pKa = 4.38RR175 pKa = 11.84TEE177 pKa = 4.4TSCPAA182 pKa = 3.7
MM1 pKa = 7.48LRR3 pKa = 11.84TSAYY7 pKa = 10.12RR8 pKa = 11.84IYY10 pKa = 10.42TRR12 pKa = 11.84PTTAITLIPTLFISIRR28 pKa = 11.84IKK30 pKa = 10.8CLTNFRR36 pKa = 11.84NAVIAFMLEE45 pKa = 3.66RR46 pKa = 11.84RR47 pKa = 11.84KK48 pKa = 10.38LIPVGAGRR56 pKa = 11.84TLVTHH61 pKa = 7.11RR62 pKa = 11.84GFPRR66 pKa = 11.84QRR68 pKa = 11.84PVLPHH73 pKa = 7.5RR74 pKa = 11.84ILQWQLTLTKK84 pKa = 9.82LLPHH88 pKa = 6.45ATINVPDD95 pKa = 3.74NRR97 pKa = 11.84ITANLALAQEE107 pKa = 4.67TPYY110 pKa = 10.93SRR112 pKa = 11.84SQARR116 pKa = 11.84SEE118 pKa = 4.05TGIPLAHH125 pKa = 5.84RR126 pKa = 11.84TVVKK130 pKa = 9.41PHH132 pKa = 4.98QEE134 pKa = 3.74RR135 pKa = 11.84YY136 pKa = 7.04STHH139 pKa = 5.49IHH141 pKa = 5.37TQAQDD146 pKa = 3.27TSRR149 pKa = 11.84GPTGGLLSGVLWVGGFVGGNPHH171 pKa = 4.5QQEE174 pKa = 4.38RR175 pKa = 11.84TEE177 pKa = 4.4TSCPAA182 pKa = 3.7
Molecular weight: 20.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2525 |
115 |
725 |
315.6 |
34.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.139 ± 0.696 | 1.267 ± 0.529 |
8.436 ± 0.904 | 8.396 ± 0.799 |
3.168 ± 0.642 | 7.921 ± 0.56 |
1.782 ± 0.49 | 3.406 ± 0.626 |
2.614 ± 0.257 | 7.802 ± 1.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.178 ± 0.206 | 3.05 ± 0.594 |
4.554 ± 0.681 | 3.01 ± 0.636 |
5.822 ± 1.172 | 7.921 ± 0.743 |
7.089 ± 0.977 | 7.881 ± 0.894 |
1.109 ± 0.236 | 2.455 ± 0.604 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
