
BtNv-AlphaCoV/SC2013
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; Nyctacovirus; Nyctalus velutinus alphacoronavirus SC-2013
Average proteome isoelectric point is 7.41
Get precalculated fractions of proteins
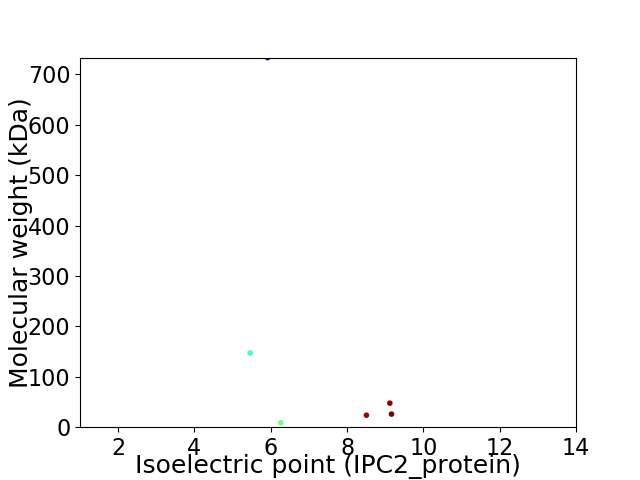
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
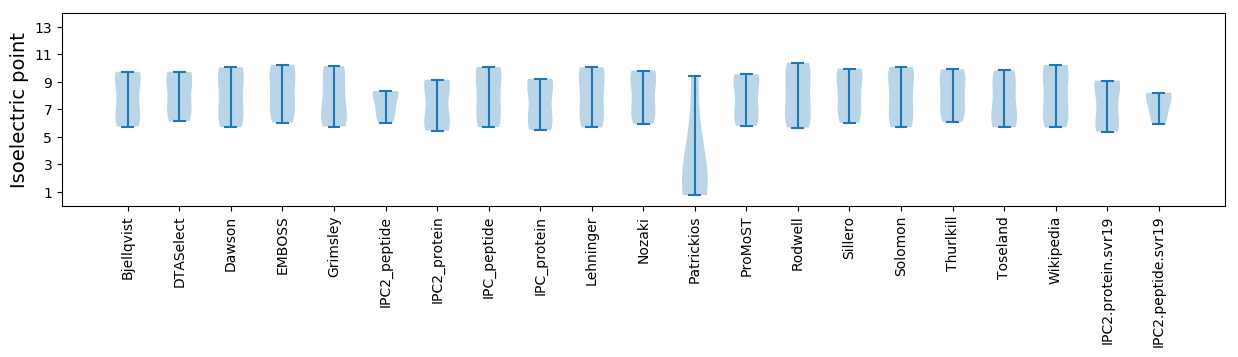
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U1WHF2|A0A0U1WHF2_9ALPC Membrane protein OS=BtNv-AlphaCoV/SC2013 OX=1503291 GN=M PE=3 SV=1
MM1 pKa = 7.78KK2 pKa = 10.1INLGLPPSTSSFVSGYY18 pKa = 10.85LPTPGNWTCRR28 pKa = 11.84NGGTTKK34 pKa = 10.76LQGHH38 pKa = 6.65ARR40 pKa = 11.84AVFMRR45 pKa = 11.84YY46 pKa = 8.29YY47 pKa = 10.57AYY49 pKa = 10.61ARR51 pKa = 11.84AWSFGVGPSTTIDD64 pKa = 3.22GHH66 pKa = 6.16YY67 pKa = 10.63GLYY70 pKa = 9.68VWHH73 pKa = 6.99SNSAGHH79 pKa = 4.81MTVRR83 pKa = 11.84ICRR86 pKa = 11.84WYY88 pKa = 10.36QDD90 pKa = 3.88RR91 pKa = 11.84DD92 pKa = 3.98PIAQNSPHH100 pKa = 6.99DD101 pKa = 4.18TTSSDD106 pKa = 3.6CILNVRR112 pKa = 11.84QPYY115 pKa = 9.54VFTHH119 pKa = 6.91AGFQVIGVSWTGEE132 pKa = 3.68AVTVYY137 pKa = 10.27GKK139 pKa = 10.63SKK141 pKa = 10.55LYY143 pKa = 10.24RR144 pKa = 11.84LYY146 pKa = 11.28VPGASLWNSVAFTCSKK162 pKa = 10.9DD163 pKa = 3.62DD164 pKa = 3.61SCGHH168 pKa = 5.98QIITKK173 pKa = 9.35PITVNATTDD182 pKa = 3.47DD183 pKa = 3.7KK184 pKa = 11.83GVITSYY190 pKa = 8.59TVCDD194 pKa = 3.49RR195 pKa = 11.84CDD197 pKa = 3.45GFPHH201 pKa = 6.72HH202 pKa = 6.86VFAVQEE208 pKa = 4.26GGKK211 pKa = 9.69IPGSFDD217 pKa = 3.13FTNWFYY223 pKa = 10.92LTNTSSPFDD232 pKa = 3.35GRR234 pKa = 11.84FVSNQPLQIQCLWPIPALTSTTGIIYY260 pKa = 9.97FNSSRR265 pKa = 11.84FTDD268 pKa = 3.11PHH270 pKa = 5.65QRR272 pKa = 11.84CNGYY276 pKa = 10.2NEE278 pKa = 4.11VGGLADD284 pKa = 3.87HH285 pKa = 6.86LRR287 pKa = 11.84FAINVTDD294 pKa = 4.3RR295 pKa = 11.84GAFQLGVISLLAVHH309 pKa = 6.45NAYY312 pKa = 10.43NFSCSNISNFVDD324 pKa = 3.11ATTIGVPFGKK334 pKa = 8.9TYY336 pKa = 10.21QPYY339 pKa = 8.43YY340 pKa = 10.61CFVTEE345 pKa = 5.37GYY347 pKa = 10.21NISANRR353 pKa = 11.84TFVGVMPSDD362 pKa = 3.33VRR364 pKa = 11.84EE365 pKa = 4.22IVVSRR370 pKa = 11.84YY371 pKa = 9.41GSVYY375 pKa = 9.83INGYY379 pKa = 10.08KK380 pKa = 9.84IFNVGEE386 pKa = 4.09LYY388 pKa = 10.95GVVLNFSSLTGSDD401 pKa = 3.63FWTVAYY407 pKa = 10.32ANEE410 pKa = 4.18VNVLVDD416 pKa = 3.32IEE418 pKa = 4.33EE419 pKa = 4.44TYY421 pKa = 9.3ITGILYY427 pKa = 10.44CDD429 pKa = 3.55TPLNRR434 pKa = 11.84LKK436 pKa = 10.75CQQQRR441 pKa = 11.84FFMDD445 pKa = 3.65DD446 pKa = 3.2GFYY449 pKa = 10.89SAIDD453 pKa = 3.5LAPPVVQTIVLLPEE467 pKa = 3.89YY468 pKa = 10.05TGLTNITLDD477 pKa = 3.5VNVTFNPPSCIQCAPTVNSILLNGEE502 pKa = 4.08SGGSVCVSTNRR513 pKa = 11.84FSVDD517 pKa = 3.51FKK519 pKa = 11.11LTVNSQAYY527 pKa = 10.03DD528 pKa = 2.96IGVRR532 pKa = 11.84TGSCPFSYY540 pKa = 10.85GSLNNFVKK548 pKa = 10.71FGSICFSLVDD558 pKa = 3.98NGGCPMPINAINYY571 pKa = 8.97IKK573 pKa = 10.0IEE575 pKa = 4.01YY576 pKa = 8.66PIGVLYY582 pKa = 8.93VTHH585 pKa = 6.75SPGEE589 pKa = 4.41AITGVPKK596 pKa = 10.97GFIKK600 pKa = 10.83RR601 pKa = 11.84LGFLDD606 pKa = 4.89ASILHH611 pKa = 6.46LNVCTEE617 pKa = 3.72YY618 pKa = 10.96NIYY621 pKa = 10.75GIVGKK626 pKa = 10.71GIINKK631 pKa = 9.66ANSTLHH637 pKa = 5.56TGIVYY642 pKa = 9.8TDD644 pKa = 3.34TAGVLVSFKK653 pKa = 11.02NVTTGDD659 pKa = 3.48IYY661 pKa = 11.4SVIPCQTSMQYY672 pKa = 11.35AVIADD677 pKa = 4.05NIVGVISADD686 pKa = 3.3TATGITFNHH695 pKa = 7.09TIATPMFYY703 pKa = 10.83YY704 pKa = 9.65STNTDD709 pKa = 3.65RR710 pKa = 11.84NCTEE714 pKa = 3.8PVLTYY719 pKa = 10.29ATMGICADD727 pKa = 3.7GAIGYY732 pKa = 7.2VQPRR736 pKa = 11.84VVSTTPATPITTGNLTIPVNFTVSIQAEE764 pKa = 4.38YY765 pKa = 10.38VQVSLRR771 pKa = 11.84AVVVDD776 pKa = 3.84CATYY780 pKa = 9.6VCNGNVRR787 pKa = 11.84CLQLLRR793 pKa = 11.84QYY795 pKa = 8.82VTACTSVEE803 pKa = 3.79NALALNARR811 pKa = 11.84LEE813 pKa = 4.34SQEE816 pKa = 4.09VADD819 pKa = 4.71MLAVDD824 pKa = 3.84YY825 pKa = 10.79AAYY828 pKa = 10.22RR829 pKa = 11.84SSLEE833 pKa = 3.88LNTPQFEE840 pKa = 4.46NGFNISAVLPAGEE853 pKa = 4.51GKK855 pKa = 10.84GSFIEE860 pKa = 5.32DD861 pKa = 3.63LLFDD865 pKa = 3.9KK866 pKa = 11.22VITNGLGTVDD876 pKa = 4.6ADD878 pKa = 3.57YY879 pKa = 11.06KK880 pKa = 10.91RR881 pKa = 11.84CIEE884 pKa = 4.21EE885 pKa = 3.81KK886 pKa = 10.71GAIADD891 pKa = 3.94VVCRR895 pKa = 11.84QYY897 pKa = 11.77YY898 pKa = 10.12KK899 pKa = 10.81GISVLPALTDD909 pKa = 3.65PARR912 pKa = 11.84MGLYY916 pKa = 9.94SASLMGALTLGAFGGGAVAAPFSIAVFSKK945 pKa = 10.78LNYY948 pKa = 9.39IALQTDD954 pKa = 5.27LIQEE958 pKa = 4.1NQKK961 pKa = 10.71LISAAFNNAMGNITKK976 pKa = 10.55AFTDD980 pKa = 3.55VNTALQHH987 pKa = 6.02VSDD990 pKa = 4.04AVKK993 pKa = 9.61TVATALNKK1001 pKa = 9.86VQDD1004 pKa = 3.84AVNTQGEE1011 pKa = 4.55ALQKK1015 pKa = 9.77LTSQLAQNFDD1025 pKa = 4.76AISSSIDD1032 pKa = 3.65DD1033 pKa = 4.42IYY1035 pKa = 11.93NKK1037 pKa = 10.43LDD1039 pKa = 3.33VLAADD1044 pKa = 3.83AQVDD1048 pKa = 3.7RR1049 pKa = 11.84LINGRR1054 pKa = 11.84LSALSTFVSAQLVKK1068 pKa = 10.62YY1069 pKa = 10.37SEE1071 pKa = 4.51VKK1073 pKa = 10.49ASRR1076 pKa = 11.84NLAMQKK1082 pKa = 10.19VNEE1085 pKa = 4.39CVKK1088 pKa = 10.43SQSSRR1093 pKa = 11.84LGFCGNGTHH1102 pKa = 7.02LFSMVTGAPDD1112 pKa = 3.52GLMFLHH1118 pKa = 6.3TVLLPTEE1125 pKa = 4.26YY1126 pKa = 10.87KK1127 pKa = 10.4EE1128 pKa = 3.97VAAWAGLCVGGKK1140 pKa = 10.25AFVLRR1145 pKa = 11.84DD1146 pKa = 3.52VQLLLFIRR1154 pKa = 11.84IIQYY1158 pKa = 10.87LVTSRR1163 pKa = 11.84NMYY1166 pKa = 9.15QPRR1169 pKa = 11.84VPQMSDD1175 pKa = 2.95FVQIQSCAITYY1186 pKa = 10.33VNLTSEE1192 pKa = 4.64EE1193 pKa = 4.19FNSVVPDD1200 pKa = 3.91YY1201 pKa = 11.33IDD1203 pKa = 3.42VNKK1206 pKa = 9.6TLEE1209 pKa = 4.48DD1210 pKa = 4.0FAATLPNRR1218 pKa = 11.84TYY1220 pKa = 11.2PDD1222 pKa = 3.84FSLDD1226 pKa = 4.0QFNHH1230 pKa = 5.93TYY1232 pKa = 11.21LNISGQISVLEE1243 pKa = 4.11NKK1245 pKa = 9.97SAEE1248 pKa = 4.06LLLITEE1254 pKa = 5.3RR1255 pKa = 11.84LQQHH1259 pKa = 5.86IQNINNSLIDD1269 pKa = 4.32LEE1271 pKa = 4.24WLDD1274 pKa = 4.42RR1275 pKa = 11.84LEE1277 pKa = 5.26TYY1279 pKa = 10.02VKK1281 pKa = 9.41WPWWVWLCFAVVFVILLGLMLWCCIATGCCGCCSCITASCAGCCDD1326 pKa = 3.47CRR1328 pKa = 11.84GKK1330 pKa = 10.56RR1331 pKa = 11.84LQRR1334 pKa = 11.84YY1335 pKa = 6.61EE1336 pKa = 3.78VEE1338 pKa = 4.83KK1339 pKa = 10.91IHH1341 pKa = 6.56IQQ1343 pKa = 3.3
MM1 pKa = 7.78KK2 pKa = 10.1INLGLPPSTSSFVSGYY18 pKa = 10.85LPTPGNWTCRR28 pKa = 11.84NGGTTKK34 pKa = 10.76LQGHH38 pKa = 6.65ARR40 pKa = 11.84AVFMRR45 pKa = 11.84YY46 pKa = 8.29YY47 pKa = 10.57AYY49 pKa = 10.61ARR51 pKa = 11.84AWSFGVGPSTTIDD64 pKa = 3.22GHH66 pKa = 6.16YY67 pKa = 10.63GLYY70 pKa = 9.68VWHH73 pKa = 6.99SNSAGHH79 pKa = 4.81MTVRR83 pKa = 11.84ICRR86 pKa = 11.84WYY88 pKa = 10.36QDD90 pKa = 3.88RR91 pKa = 11.84DD92 pKa = 3.98PIAQNSPHH100 pKa = 6.99DD101 pKa = 4.18TTSSDD106 pKa = 3.6CILNVRR112 pKa = 11.84QPYY115 pKa = 9.54VFTHH119 pKa = 6.91AGFQVIGVSWTGEE132 pKa = 3.68AVTVYY137 pKa = 10.27GKK139 pKa = 10.63SKK141 pKa = 10.55LYY143 pKa = 10.24RR144 pKa = 11.84LYY146 pKa = 11.28VPGASLWNSVAFTCSKK162 pKa = 10.9DD163 pKa = 3.62DD164 pKa = 3.61SCGHH168 pKa = 5.98QIITKK173 pKa = 9.35PITVNATTDD182 pKa = 3.47DD183 pKa = 3.7KK184 pKa = 11.83GVITSYY190 pKa = 8.59TVCDD194 pKa = 3.49RR195 pKa = 11.84CDD197 pKa = 3.45GFPHH201 pKa = 6.72HH202 pKa = 6.86VFAVQEE208 pKa = 4.26GGKK211 pKa = 9.69IPGSFDD217 pKa = 3.13FTNWFYY223 pKa = 10.92LTNTSSPFDD232 pKa = 3.35GRR234 pKa = 11.84FVSNQPLQIQCLWPIPALTSTTGIIYY260 pKa = 9.97FNSSRR265 pKa = 11.84FTDD268 pKa = 3.11PHH270 pKa = 5.65QRR272 pKa = 11.84CNGYY276 pKa = 10.2NEE278 pKa = 4.11VGGLADD284 pKa = 3.87HH285 pKa = 6.86LRR287 pKa = 11.84FAINVTDD294 pKa = 4.3RR295 pKa = 11.84GAFQLGVISLLAVHH309 pKa = 6.45NAYY312 pKa = 10.43NFSCSNISNFVDD324 pKa = 3.11ATTIGVPFGKK334 pKa = 8.9TYY336 pKa = 10.21QPYY339 pKa = 8.43YY340 pKa = 10.61CFVTEE345 pKa = 5.37GYY347 pKa = 10.21NISANRR353 pKa = 11.84TFVGVMPSDD362 pKa = 3.33VRR364 pKa = 11.84EE365 pKa = 4.22IVVSRR370 pKa = 11.84YY371 pKa = 9.41GSVYY375 pKa = 9.83INGYY379 pKa = 10.08KK380 pKa = 9.84IFNVGEE386 pKa = 4.09LYY388 pKa = 10.95GVVLNFSSLTGSDD401 pKa = 3.63FWTVAYY407 pKa = 10.32ANEE410 pKa = 4.18VNVLVDD416 pKa = 3.32IEE418 pKa = 4.33EE419 pKa = 4.44TYY421 pKa = 9.3ITGILYY427 pKa = 10.44CDD429 pKa = 3.55TPLNRR434 pKa = 11.84LKK436 pKa = 10.75CQQQRR441 pKa = 11.84FFMDD445 pKa = 3.65DD446 pKa = 3.2GFYY449 pKa = 10.89SAIDD453 pKa = 3.5LAPPVVQTIVLLPEE467 pKa = 3.89YY468 pKa = 10.05TGLTNITLDD477 pKa = 3.5VNVTFNPPSCIQCAPTVNSILLNGEE502 pKa = 4.08SGGSVCVSTNRR513 pKa = 11.84FSVDD517 pKa = 3.51FKK519 pKa = 11.11LTVNSQAYY527 pKa = 10.03DD528 pKa = 2.96IGVRR532 pKa = 11.84TGSCPFSYY540 pKa = 10.85GSLNNFVKK548 pKa = 10.71FGSICFSLVDD558 pKa = 3.98NGGCPMPINAINYY571 pKa = 8.97IKK573 pKa = 10.0IEE575 pKa = 4.01YY576 pKa = 8.66PIGVLYY582 pKa = 8.93VTHH585 pKa = 6.75SPGEE589 pKa = 4.41AITGVPKK596 pKa = 10.97GFIKK600 pKa = 10.83RR601 pKa = 11.84LGFLDD606 pKa = 4.89ASILHH611 pKa = 6.46LNVCTEE617 pKa = 3.72YY618 pKa = 10.96NIYY621 pKa = 10.75GIVGKK626 pKa = 10.71GIINKK631 pKa = 9.66ANSTLHH637 pKa = 5.56TGIVYY642 pKa = 9.8TDD644 pKa = 3.34TAGVLVSFKK653 pKa = 11.02NVTTGDD659 pKa = 3.48IYY661 pKa = 11.4SVIPCQTSMQYY672 pKa = 11.35AVIADD677 pKa = 4.05NIVGVISADD686 pKa = 3.3TATGITFNHH695 pKa = 7.09TIATPMFYY703 pKa = 10.83YY704 pKa = 9.65STNTDD709 pKa = 3.65RR710 pKa = 11.84NCTEE714 pKa = 3.8PVLTYY719 pKa = 10.29ATMGICADD727 pKa = 3.7GAIGYY732 pKa = 7.2VQPRR736 pKa = 11.84VVSTTPATPITTGNLTIPVNFTVSIQAEE764 pKa = 4.38YY765 pKa = 10.38VQVSLRR771 pKa = 11.84AVVVDD776 pKa = 3.84CATYY780 pKa = 9.6VCNGNVRR787 pKa = 11.84CLQLLRR793 pKa = 11.84QYY795 pKa = 8.82VTACTSVEE803 pKa = 3.79NALALNARR811 pKa = 11.84LEE813 pKa = 4.34SQEE816 pKa = 4.09VADD819 pKa = 4.71MLAVDD824 pKa = 3.84YY825 pKa = 10.79AAYY828 pKa = 10.22RR829 pKa = 11.84SSLEE833 pKa = 3.88LNTPQFEE840 pKa = 4.46NGFNISAVLPAGEE853 pKa = 4.51GKK855 pKa = 10.84GSFIEE860 pKa = 5.32DD861 pKa = 3.63LLFDD865 pKa = 3.9KK866 pKa = 11.22VITNGLGTVDD876 pKa = 4.6ADD878 pKa = 3.57YY879 pKa = 11.06KK880 pKa = 10.91RR881 pKa = 11.84CIEE884 pKa = 4.21EE885 pKa = 3.81KK886 pKa = 10.71GAIADD891 pKa = 3.94VVCRR895 pKa = 11.84QYY897 pKa = 11.77YY898 pKa = 10.12KK899 pKa = 10.81GISVLPALTDD909 pKa = 3.65PARR912 pKa = 11.84MGLYY916 pKa = 9.94SASLMGALTLGAFGGGAVAAPFSIAVFSKK945 pKa = 10.78LNYY948 pKa = 9.39IALQTDD954 pKa = 5.27LIQEE958 pKa = 4.1NQKK961 pKa = 10.71LISAAFNNAMGNITKK976 pKa = 10.55AFTDD980 pKa = 3.55VNTALQHH987 pKa = 6.02VSDD990 pKa = 4.04AVKK993 pKa = 9.61TVATALNKK1001 pKa = 9.86VQDD1004 pKa = 3.84AVNTQGEE1011 pKa = 4.55ALQKK1015 pKa = 9.77LTSQLAQNFDD1025 pKa = 4.76AISSSIDD1032 pKa = 3.65DD1033 pKa = 4.42IYY1035 pKa = 11.93NKK1037 pKa = 10.43LDD1039 pKa = 3.33VLAADD1044 pKa = 3.83AQVDD1048 pKa = 3.7RR1049 pKa = 11.84LINGRR1054 pKa = 11.84LSALSTFVSAQLVKK1068 pKa = 10.62YY1069 pKa = 10.37SEE1071 pKa = 4.51VKK1073 pKa = 10.49ASRR1076 pKa = 11.84NLAMQKK1082 pKa = 10.19VNEE1085 pKa = 4.39CVKK1088 pKa = 10.43SQSSRR1093 pKa = 11.84LGFCGNGTHH1102 pKa = 7.02LFSMVTGAPDD1112 pKa = 3.52GLMFLHH1118 pKa = 6.3TVLLPTEE1125 pKa = 4.26YY1126 pKa = 10.87KK1127 pKa = 10.4EE1128 pKa = 3.97VAAWAGLCVGGKK1140 pKa = 10.25AFVLRR1145 pKa = 11.84DD1146 pKa = 3.52VQLLLFIRR1154 pKa = 11.84IIQYY1158 pKa = 10.87LVTSRR1163 pKa = 11.84NMYY1166 pKa = 9.15QPRR1169 pKa = 11.84VPQMSDD1175 pKa = 2.95FVQIQSCAITYY1186 pKa = 10.33VNLTSEE1192 pKa = 4.64EE1193 pKa = 4.19FNSVVPDD1200 pKa = 3.91YY1201 pKa = 11.33IDD1203 pKa = 3.42VNKK1206 pKa = 9.6TLEE1209 pKa = 4.48DD1210 pKa = 4.0FAATLPNRR1218 pKa = 11.84TYY1220 pKa = 11.2PDD1222 pKa = 3.84FSLDD1226 pKa = 4.0QFNHH1230 pKa = 5.93TYY1232 pKa = 11.21LNISGQISVLEE1243 pKa = 4.11NKK1245 pKa = 9.97SAEE1248 pKa = 4.06LLLITEE1254 pKa = 5.3RR1255 pKa = 11.84LQQHH1259 pKa = 5.86IQNINNSLIDD1269 pKa = 4.32LEE1271 pKa = 4.24WLDD1274 pKa = 4.42RR1275 pKa = 11.84LEE1277 pKa = 5.26TYY1279 pKa = 10.02VKK1281 pKa = 9.41WPWWVWLCFAVVFVILLGLMLWCCIATGCCGCCSCITASCAGCCDD1326 pKa = 3.47CRR1328 pKa = 11.84GKK1330 pKa = 10.56RR1331 pKa = 11.84LQRR1334 pKa = 11.84YY1335 pKa = 6.61EE1336 pKa = 3.78VEE1338 pKa = 4.83KK1339 pKa = 10.91IHH1341 pKa = 6.56IQQ1343 pKa = 3.3
Molecular weight: 147.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U1WHG1|A0A0U1WHG1_9ALPC 3C-like proteinase OS=BtNv-AlphaCoV/SC2013 OX=1503291 PE=3 SV=1
MM1 pKa = 7.5SSSKK5 pKa = 11.24GNVGFDD11 pKa = 2.82NAARR15 pKa = 11.84GRR17 pKa = 11.84SGRR20 pKa = 11.84VPFSFYY26 pKa = 10.58MPVINNSSQPFYY38 pKa = 11.42KK39 pKa = 10.64VMPQNAVPKK48 pKa = 10.12GQGNKK53 pKa = 9.62DD54 pKa = 3.41QQIGYY59 pKa = 9.11WNEE62 pKa = 3.54QVRR65 pKa = 11.84WRR67 pKa = 11.84MVKK70 pKa = 8.84GTRR73 pKa = 11.84KK74 pKa = 9.86DD75 pKa = 4.25LPSKK79 pKa = 8.43WHH81 pKa = 6.58FYY83 pKa = 10.71YY84 pKa = 10.91LGTGPHH90 pKa = 6.95ADD92 pKa = 2.84LKK94 pKa = 10.73FRR96 pKa = 11.84QRR98 pKa = 11.84QQGVFWVAKK107 pKa = 9.07EE108 pKa = 3.93GAKK111 pKa = 10.44AEE113 pKa = 4.07PTGLGTRR120 pKa = 11.84GRR122 pKa = 11.84NAEE125 pKa = 3.99LTTPIFNPGLPDD137 pKa = 3.77SIEE140 pKa = 4.01IVDD143 pKa = 3.7QYY145 pKa = 11.8SRR147 pKa = 11.84PNSRR151 pKa = 11.84ASSRR155 pKa = 11.84ARR157 pKa = 11.84SQSNDD162 pKa = 2.8QGNRR166 pKa = 11.84SRR168 pKa = 11.84SQSNNRR174 pKa = 11.84AQSNNRR180 pKa = 11.84SQSRR184 pKa = 11.84GRR186 pKa = 11.84QNQNQNNQPTGDD198 pKa = 3.75GGSNGQRR205 pKa = 11.84NQPRR209 pKa = 11.84NRR211 pKa = 11.84SNSRR215 pKa = 11.84NRR217 pKa = 11.84SGNQGRR223 pKa = 11.84NGGSQQDD230 pKa = 3.39LVAAVRR236 pKa = 11.84EE237 pKa = 4.29ALAGMGFKK245 pKa = 10.69PNTSGSGRR253 pKa = 11.84NTPVKK258 pKa = 10.27VPKK261 pKa = 10.25GDD263 pKa = 3.51KK264 pKa = 9.99PLNKK268 pKa = 8.24PTKK271 pKa = 9.61APASQVEE278 pKa = 4.24KK279 pKa = 9.98PVWKK283 pKa = 8.87RR284 pKa = 11.84TPHH287 pKa = 4.91SQEE290 pKa = 3.68NVEE293 pKa = 4.42VCFGPRR299 pKa = 11.84DD300 pKa = 3.64TYY302 pKa = 11.68QNFGDD307 pKa = 4.02SQLVRR312 pKa = 11.84LGVDD316 pKa = 3.37YY317 pKa = 10.33PHH319 pKa = 6.77YY320 pKa = 9.41PQIGEE325 pKa = 5.1LIPSQAALLFGSEE338 pKa = 3.81ITAHH342 pKa = 5.69EE343 pKa = 4.6RR344 pKa = 11.84GDD346 pKa = 4.15NIQLTYY352 pKa = 9.36IYY354 pKa = 10.7KK355 pKa = 8.98MEE357 pKa = 4.0VPKK360 pKa = 10.67DD361 pKa = 3.88HH362 pKa = 7.17KK363 pKa = 11.01SLAAFLPHH371 pKa = 6.55IGAYY375 pKa = 9.99ADD377 pKa = 3.5STEE380 pKa = 5.07DD381 pKa = 3.27VTLPPALPPKK391 pKa = 7.99QQRR394 pKa = 11.84LRR396 pKa = 11.84RR397 pKa = 11.84SASTEE402 pKa = 3.74VLADD406 pKa = 3.48TTSVVDD412 pKa = 3.63EE413 pKa = 4.71EE414 pKa = 4.44EE415 pKa = 4.32VEE417 pKa = 4.36EE418 pKa = 4.5VVDD421 pKa = 5.25DD422 pKa = 3.74VTGQDD427 pKa = 3.01EE428 pKa = 4.52TFAA431 pKa = 5.66
MM1 pKa = 7.5SSSKK5 pKa = 11.24GNVGFDD11 pKa = 2.82NAARR15 pKa = 11.84GRR17 pKa = 11.84SGRR20 pKa = 11.84VPFSFYY26 pKa = 10.58MPVINNSSQPFYY38 pKa = 11.42KK39 pKa = 10.64VMPQNAVPKK48 pKa = 10.12GQGNKK53 pKa = 9.62DD54 pKa = 3.41QQIGYY59 pKa = 9.11WNEE62 pKa = 3.54QVRR65 pKa = 11.84WRR67 pKa = 11.84MVKK70 pKa = 8.84GTRR73 pKa = 11.84KK74 pKa = 9.86DD75 pKa = 4.25LPSKK79 pKa = 8.43WHH81 pKa = 6.58FYY83 pKa = 10.71YY84 pKa = 10.91LGTGPHH90 pKa = 6.95ADD92 pKa = 2.84LKK94 pKa = 10.73FRR96 pKa = 11.84QRR98 pKa = 11.84QQGVFWVAKK107 pKa = 9.07EE108 pKa = 3.93GAKK111 pKa = 10.44AEE113 pKa = 4.07PTGLGTRR120 pKa = 11.84GRR122 pKa = 11.84NAEE125 pKa = 3.99LTTPIFNPGLPDD137 pKa = 3.77SIEE140 pKa = 4.01IVDD143 pKa = 3.7QYY145 pKa = 11.8SRR147 pKa = 11.84PNSRR151 pKa = 11.84ASSRR155 pKa = 11.84ARR157 pKa = 11.84SQSNDD162 pKa = 2.8QGNRR166 pKa = 11.84SRR168 pKa = 11.84SQSNNRR174 pKa = 11.84AQSNNRR180 pKa = 11.84SQSRR184 pKa = 11.84GRR186 pKa = 11.84QNQNQNNQPTGDD198 pKa = 3.75GGSNGQRR205 pKa = 11.84NQPRR209 pKa = 11.84NRR211 pKa = 11.84SNSRR215 pKa = 11.84NRR217 pKa = 11.84SGNQGRR223 pKa = 11.84NGGSQQDD230 pKa = 3.39LVAAVRR236 pKa = 11.84EE237 pKa = 4.29ALAGMGFKK245 pKa = 10.69PNTSGSGRR253 pKa = 11.84NTPVKK258 pKa = 10.27VPKK261 pKa = 10.25GDD263 pKa = 3.51KK264 pKa = 9.99PLNKK268 pKa = 8.24PTKK271 pKa = 9.61APASQVEE278 pKa = 4.24KK279 pKa = 9.98PVWKK283 pKa = 8.87RR284 pKa = 11.84TPHH287 pKa = 4.91SQEE290 pKa = 3.68NVEE293 pKa = 4.42VCFGPRR299 pKa = 11.84DD300 pKa = 3.64TYY302 pKa = 11.68QNFGDD307 pKa = 4.02SQLVRR312 pKa = 11.84LGVDD316 pKa = 3.37YY317 pKa = 10.33PHH319 pKa = 6.77YY320 pKa = 9.41PQIGEE325 pKa = 5.1LIPSQAALLFGSEE338 pKa = 3.81ITAHH342 pKa = 5.69EE343 pKa = 4.6RR344 pKa = 11.84GDD346 pKa = 4.15NIQLTYY352 pKa = 9.36IYY354 pKa = 10.7KK355 pKa = 8.98MEE357 pKa = 4.0VPKK360 pKa = 10.67DD361 pKa = 3.88HH362 pKa = 7.17KK363 pKa = 11.01SLAAFLPHH371 pKa = 6.55IGAYY375 pKa = 9.99ADD377 pKa = 3.5STEE380 pKa = 5.07DD381 pKa = 3.27VTLPPALPPKK391 pKa = 7.99QQRR394 pKa = 11.84LRR396 pKa = 11.84RR397 pKa = 11.84SASTEE402 pKa = 3.74VLADD406 pKa = 3.48TTSVVDD412 pKa = 3.63EE413 pKa = 4.71EE414 pKa = 4.44EE415 pKa = 4.32VEE417 pKa = 4.36EE418 pKa = 4.5VVDD421 pKa = 5.25DD422 pKa = 3.74VTGQDD427 pKa = 3.01EE428 pKa = 4.52TFAA431 pKa = 5.66
Molecular weight: 47.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8882 |
75 |
6598 |
1480.3 |
164.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.71 ± 0.244 | 3.445 ± 0.8 |
5.562 ± 0.68 | 3.67 ± 0.431 |
5.663 ± 0.641 | 6.834 ± 0.433 |
1.959 ± 0.149 | 4.447 ± 0.918 |
4.976 ± 0.854 | 8.759 ± 0.598 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.027 ± 0.288 | 5.506 ± 0.504 |
3.637 ± 0.628 | 2.984 ± 0.874 |
3.839 ± 0.807 | 7.329 ± 0.324 |
6.192 ± 0.589 | 10.493 ± 0.722 |
1.317 ± 0.266 | 4.65 ± 0.324 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
