
Human associated cyclovirus 1 (isolate Homo sapiens/Pakistan/PK5510/2007) (HuCyV-1) (Cyclovirus PK5510)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Cyclovirus; Human associated cyclovirus 1
Average proteome isoelectric point is 8.6
Get precalculated fractions of proteins
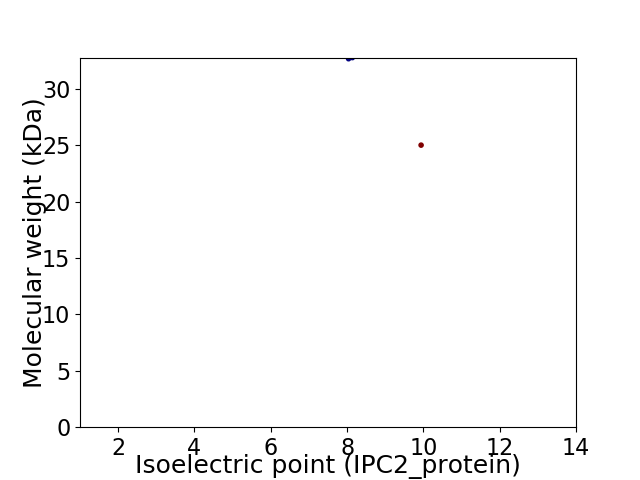
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
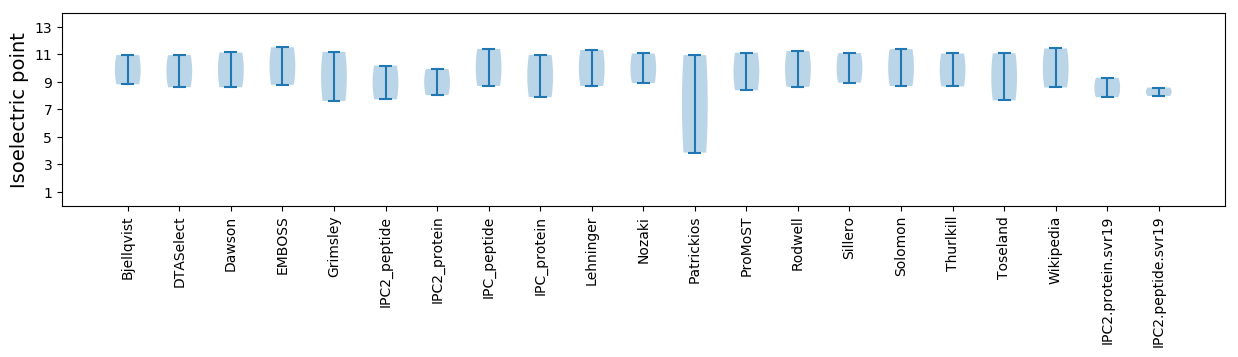
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|D4N3P3|CAPSD_HCYV5 Capsid protein OS=Human associated cyclovirus 1 (isolate Homo sapiens/Pakistan/PK5510/2007) OX=742918 GN=Cap PE=3 SV=1
MM1 pKa = 7.38SNSTVRR7 pKa = 11.84RR8 pKa = 11.84FCFTWNNYY16 pKa = 8.05TEE18 pKa = 5.06LNYY21 pKa = 10.86ALCQEE26 pKa = 5.14FIKK29 pKa = 10.42KK30 pKa = 7.9YY31 pKa = 9.24CKK33 pKa = 10.01YY34 pKa = 10.37GIVGKK39 pKa = 10.07EE40 pKa = 3.76LAPTTNTPHH49 pKa = 6.74LQGFCNLQKK58 pKa = 10.56PMRR61 pKa = 11.84FSTIKK66 pKa = 10.18KK67 pKa = 9.42RR68 pKa = 11.84LDD70 pKa = 3.25NGIHH74 pKa = 6.27IEE76 pKa = 3.9KK77 pKa = 10.89SMGSDD82 pKa = 3.28TQNQTYY88 pKa = 9.73CSKK91 pKa = 10.76SGEE94 pKa = 4.14FFEE97 pKa = 6.59AGDD100 pKa = 3.94PQCQGKK106 pKa = 10.38RR107 pKa = 11.84NDD109 pKa = 3.68LQSVVDD115 pKa = 4.81TIQAGNGSLSSIANEE130 pKa = 4.23HH131 pKa = 5.04PTAYY135 pKa = 9.28IRR137 pKa = 11.84YY138 pKa = 7.99FRR140 pKa = 11.84GIQEE144 pKa = 4.25YY145 pKa = 9.66IKK147 pKa = 9.02TVRR150 pKa = 11.84PIPPRR155 pKa = 11.84YY156 pKa = 9.07HH157 pKa = 5.28KK158 pKa = 9.86TEE160 pKa = 3.2VRR162 pKa = 11.84YY163 pKa = 8.98YY164 pKa = 10.09HH165 pKa = 6.81GPPGSGKK172 pKa = 8.61SRR174 pKa = 11.84RR175 pKa = 11.84ALEE178 pKa = 4.0EE179 pKa = 3.77ATALASDD186 pKa = 4.84LNDD189 pKa = 2.77IYY191 pKa = 11.34YY192 pKa = 10.39KK193 pKa = 10.63PRR195 pKa = 11.84GTWWDD200 pKa = 3.84GYY202 pKa = 10.05KK203 pKa = 9.86QQSCVIIDD211 pKa = 4.9DD212 pKa = 4.45FYY214 pKa = 11.76GWIKK218 pKa = 9.81YY219 pKa = 10.71DD220 pKa = 3.92EE221 pKa = 4.17MLKK224 pKa = 9.91ICDD227 pKa = 3.53RR228 pKa = 11.84YY229 pKa = 9.77PYY231 pKa = 10.1KK232 pKa = 10.94VQIKK236 pKa = 10.43GGFEE240 pKa = 4.14EE241 pKa = 4.92FTSKK245 pKa = 10.99YY246 pKa = 9.93IWITSNIDD254 pKa = 3.02TNLLYY259 pKa = 10.79KK260 pKa = 10.82FNDD263 pKa = 3.53YY264 pKa = 11.34NDD266 pKa = 3.56TAFVRR271 pKa = 11.84RR272 pKa = 11.84IEE274 pKa = 4.02IKK276 pKa = 10.79LLIEE280 pKa = 4.14
MM1 pKa = 7.38SNSTVRR7 pKa = 11.84RR8 pKa = 11.84FCFTWNNYY16 pKa = 8.05TEE18 pKa = 5.06LNYY21 pKa = 10.86ALCQEE26 pKa = 5.14FIKK29 pKa = 10.42KK30 pKa = 7.9YY31 pKa = 9.24CKK33 pKa = 10.01YY34 pKa = 10.37GIVGKK39 pKa = 10.07EE40 pKa = 3.76LAPTTNTPHH49 pKa = 6.74LQGFCNLQKK58 pKa = 10.56PMRR61 pKa = 11.84FSTIKK66 pKa = 10.18KK67 pKa = 9.42RR68 pKa = 11.84LDD70 pKa = 3.25NGIHH74 pKa = 6.27IEE76 pKa = 3.9KK77 pKa = 10.89SMGSDD82 pKa = 3.28TQNQTYY88 pKa = 9.73CSKK91 pKa = 10.76SGEE94 pKa = 4.14FFEE97 pKa = 6.59AGDD100 pKa = 3.94PQCQGKK106 pKa = 10.38RR107 pKa = 11.84NDD109 pKa = 3.68LQSVVDD115 pKa = 4.81TIQAGNGSLSSIANEE130 pKa = 4.23HH131 pKa = 5.04PTAYY135 pKa = 9.28IRR137 pKa = 11.84YY138 pKa = 7.99FRR140 pKa = 11.84GIQEE144 pKa = 4.25YY145 pKa = 9.66IKK147 pKa = 9.02TVRR150 pKa = 11.84PIPPRR155 pKa = 11.84YY156 pKa = 9.07HH157 pKa = 5.28KK158 pKa = 9.86TEE160 pKa = 3.2VRR162 pKa = 11.84YY163 pKa = 8.98YY164 pKa = 10.09HH165 pKa = 6.81GPPGSGKK172 pKa = 8.61SRR174 pKa = 11.84RR175 pKa = 11.84ALEE178 pKa = 4.0EE179 pKa = 3.77ATALASDD186 pKa = 4.84LNDD189 pKa = 2.77IYY191 pKa = 11.34YY192 pKa = 10.39KK193 pKa = 10.63PRR195 pKa = 11.84GTWWDD200 pKa = 3.84GYY202 pKa = 10.05KK203 pKa = 9.86QQSCVIIDD211 pKa = 4.9DD212 pKa = 4.45FYY214 pKa = 11.76GWIKK218 pKa = 9.81YY219 pKa = 10.71DD220 pKa = 3.92EE221 pKa = 4.17MLKK224 pKa = 9.91ICDD227 pKa = 3.53RR228 pKa = 11.84YY229 pKa = 9.77PYY231 pKa = 10.1KK232 pKa = 10.94VQIKK236 pKa = 10.43GGFEE240 pKa = 4.14EE241 pKa = 4.92FTSKK245 pKa = 10.99YY246 pKa = 9.93IWITSNIDD254 pKa = 3.02TNLLYY259 pKa = 10.79KK260 pKa = 10.82FNDD263 pKa = 3.53YY264 pKa = 11.34NDD266 pKa = 3.56TAFVRR271 pKa = 11.84RR272 pKa = 11.84IEE274 pKa = 4.02IKK276 pKa = 10.79LLIEE280 pKa = 4.14
Molecular weight: 32.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|D4N3P3|CAPSD_HCYV5 Capsid protein OS=Human associated cyclovirus 1 (isolate Homo sapiens/Pakistan/PK5510/2007) OX=742918 GN=Cap PE=3 SV=1
MM1 pKa = 7.52PFSRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84VVRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84KK14 pKa = 8.54PVRR17 pKa = 11.84RR18 pKa = 11.84LRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84FFKK28 pKa = 10.43RR29 pKa = 11.84AGRR32 pKa = 11.84GSFRR36 pKa = 11.84VKK38 pKa = 9.6LTRR41 pKa = 11.84FVSITQDD48 pKa = 2.64ISKK51 pKa = 7.04TTQFSFEE58 pKa = 4.04ITPSDD63 pKa = 3.44FTEE66 pKa = 4.42FATLADD72 pKa = 3.39AFEE75 pKa = 4.46AVRR78 pKa = 11.84FTRR81 pKa = 11.84AKK83 pKa = 9.56VTVLPQQNVSNNSTSLIPAYY103 pKa = 10.98CMFPWHH109 pKa = 6.92KK110 pKa = 9.99EE111 pKa = 3.91LPTVATFNGFLSIDD125 pKa = 3.41RR126 pKa = 11.84AKK128 pKa = 10.56CFRR131 pKa = 11.84GTQVGIQHH139 pKa = 6.07YY140 pKa = 10.21VPSTLDD146 pKa = 3.49SIQTKK151 pKa = 9.73GAAGYY156 pKa = 8.47EE157 pKa = 4.3VQSVQTKK164 pKa = 8.83YY165 pKa = 10.87KK166 pKa = 8.88PTIQISGGASTVVLYY181 pKa = 10.03TGALGMQALSDD192 pKa = 3.82APEE195 pKa = 4.19NAQAHH200 pKa = 5.09YY201 pKa = 10.25NIKK204 pKa = 9.91IDD206 pKa = 3.47MWCTFINQTSFNKK219 pKa = 10.34
MM1 pKa = 7.52PFSRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84VVRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84KK14 pKa = 8.54PVRR17 pKa = 11.84RR18 pKa = 11.84LRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84FFKK28 pKa = 10.43RR29 pKa = 11.84AGRR32 pKa = 11.84GSFRR36 pKa = 11.84VKK38 pKa = 9.6LTRR41 pKa = 11.84FVSITQDD48 pKa = 2.64ISKK51 pKa = 7.04TTQFSFEE58 pKa = 4.04ITPSDD63 pKa = 3.44FTEE66 pKa = 4.42FATLADD72 pKa = 3.39AFEE75 pKa = 4.46AVRR78 pKa = 11.84FTRR81 pKa = 11.84AKK83 pKa = 9.56VTVLPQQNVSNNSTSLIPAYY103 pKa = 10.98CMFPWHH109 pKa = 6.92KK110 pKa = 9.99EE111 pKa = 3.91LPTVATFNGFLSIDD125 pKa = 3.41RR126 pKa = 11.84AKK128 pKa = 10.56CFRR131 pKa = 11.84GTQVGIQHH139 pKa = 6.07YY140 pKa = 10.21VPSTLDD146 pKa = 3.49SIQTKK151 pKa = 9.73GAAGYY156 pKa = 8.47EE157 pKa = 4.3VQSVQTKK164 pKa = 8.83YY165 pKa = 10.87KK166 pKa = 8.88PTIQISGGASTVVLYY181 pKa = 10.03TGALGMQALSDD192 pKa = 3.82APEE195 pKa = 4.19NAQAHH200 pKa = 5.09YY201 pKa = 10.25NIKK204 pKa = 9.91IDD206 pKa = 3.47MWCTFINQTSFNKK219 pKa = 10.34
Molecular weight: 25.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
499 |
219 |
280 |
249.5 |
28.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.611 ± 1.32 | 2.204 ± 0.512 |
4.409 ± 0.744 | 4.409 ± 1.024 |
6.012 ± 1.074 | 6.012 ± 0.607 |
1.603 ± 0.143 | 7.014 ± 0.942 |
6.814 ± 0.819 | 5.411 ± 0.238 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.603 ± 0.137 | 4.81 ± 0.71 |
4.409 ± 0.097 | 5.21 ± 0.445 |
7.816 ± 1.649 | 6.814 ± 0.582 |
8.016 ± 0.965 | 5.01 ± 1.409 |
1.403 ± 0.3 | 5.411 ± 1.639 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
