
Tobacco streak virus (strain WC) (TSV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Ilarvirus; Tobacco streak virus
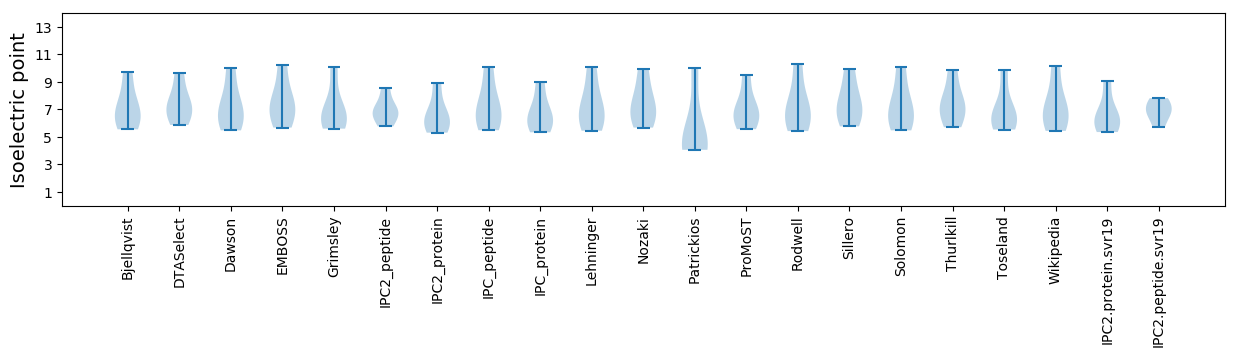
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>sp|P89679|2B_TOBSV Suppressor of silencing 2b OS=Tobacco streak virus (strain WC) OX=12318 GN=ORF2b PE=3 SV=1
MM1 pKa = 7.83DD2 pKa = 5.53LVIKK6 pKa = 10.56NLIVYY11 pKa = 8.55HH12 pKa = 5.57LRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84IDD18 pKa = 2.76IGTSFGIEE26 pKa = 3.65PADD29 pKa = 3.82YY30 pKa = 10.41IDD32 pKa = 3.67WVKK35 pKa = 10.78VFLLKK40 pKa = 10.42FIIEE44 pKa = 4.12HH45 pKa = 5.26TARR48 pKa = 11.84FADD51 pKa = 4.68FATIHH56 pKa = 5.02TTMLLVLGEE65 pKa = 4.74DD66 pKa = 3.58DD67 pKa = 3.88PNYY70 pKa = 10.36VEE72 pKa = 5.19KK73 pKa = 9.84DD74 pKa = 3.61TPIMEE79 pKa = 4.98IDD81 pKa = 4.14PFYY84 pKa = 11.2LPYY87 pKa = 10.75DD88 pKa = 4.42DD89 pKa = 6.34LDD91 pKa = 3.39VDD93 pKa = 4.07YY94 pKa = 10.73TSLRR98 pKa = 11.84VCGDD102 pKa = 3.42EE103 pKa = 5.19DD104 pKa = 3.93QSCSDD109 pKa = 4.01RR110 pKa = 11.84DD111 pKa = 3.67EE112 pKa = 5.07LSDD115 pKa = 4.08FISNISHH122 pKa = 6.27IPEE125 pKa = 4.21GTSWGSEE132 pKa = 3.59SDD134 pKa = 3.47TSFVEE139 pKa = 4.1HH140 pKa = 7.24LEE142 pKa = 4.41TIQDD146 pKa = 3.34IPTKK150 pKa = 10.76CEE152 pKa = 3.63IADD155 pKa = 3.93KK156 pKa = 10.05PVEE159 pKa = 4.77EE160 pKa = 5.36IPFDD164 pKa = 4.63DD165 pKa = 5.22DD166 pKa = 4.06GKK168 pKa = 10.75VVNDD172 pKa = 3.18VWVDD176 pKa = 3.61AEE178 pKa = 4.56LSNAPEE184 pKa = 4.1ISCDD188 pKa = 3.28ADD190 pKa = 2.89IRR192 pKa = 11.84ACGFVSLRR200 pKa = 11.84LLEE203 pKa = 4.17SSRR206 pKa = 11.84GYY208 pKa = 9.84PKK210 pKa = 8.65WTPEE214 pKa = 3.93RR215 pKa = 11.84VSSGLNPDD223 pKa = 3.99LPVNSKK229 pKa = 9.96PAVDD233 pKa = 4.71EE234 pKa = 4.31IFPHH238 pKa = 5.7HH239 pKa = 6.92HH240 pKa = 6.28SVDD243 pKa = 3.3DD244 pKa = 4.14RR245 pKa = 11.84FFQEE249 pKa = 3.7WVEE252 pKa = 3.99THH254 pKa = 7.34DD255 pKa = 3.95IDD257 pKa = 6.24LEE259 pKa = 4.47VTSCDD264 pKa = 3.8LDD266 pKa = 3.23MSTFNDD272 pKa = 3.13WTKK275 pKa = 11.11GVDD278 pKa = 3.3TRR280 pKa = 11.84LVPNMSVGGLSHH292 pKa = 7.1RR293 pKa = 11.84VPTQRR298 pKa = 11.84EE299 pKa = 3.62ALLAIKK305 pKa = 9.37KK306 pKa = 10.3RR307 pKa = 11.84NMNVPEE313 pKa = 4.11LQSNFDD319 pKa = 3.87HH320 pKa = 7.95DD321 pKa = 5.07DD322 pKa = 3.38VLNRR326 pKa = 11.84CVTRR330 pKa = 11.84FITHH334 pKa = 5.6VVDD337 pKa = 3.52KK338 pKa = 10.93TRR340 pKa = 11.84LSKK343 pKa = 10.94LNPISGEE350 pKa = 3.79EE351 pKa = 3.48LHH353 pKa = 6.79YY354 pKa = 10.77FNQYY358 pKa = 10.65LEE360 pKa = 4.47NKK362 pKa = 9.44NPPLSEE368 pKa = 4.1YY369 pKa = 10.16KK370 pKa = 10.95GPVPLVALDD379 pKa = 4.49KK380 pKa = 10.26YY381 pKa = 9.66MHH383 pKa = 6.57MIKK386 pKa = 7.75TTLKK390 pKa = 10.38PVEE393 pKa = 4.37EE394 pKa = 4.96DD395 pKa = 3.04NLHH398 pKa = 6.21IEE400 pKa = 4.23RR401 pKa = 11.84PIPATITYY409 pKa = 8.34HH410 pKa = 5.89KK411 pKa = 9.69KK412 pKa = 9.51GVVMMTSPYY421 pKa = 9.61FLCAMVRR428 pKa = 11.84LLYY431 pKa = 10.08VLKK434 pKa = 10.74SKK436 pKa = 10.49FVVPTGKK443 pKa = 9.74YY444 pKa = 9.12HH445 pKa = 7.59QIFQMNPEE453 pKa = 4.33LLKK456 pKa = 10.62HH457 pKa = 5.58SKK459 pKa = 8.92EE460 pKa = 3.97FKK462 pKa = 10.52EE463 pKa = 3.83IDD465 pKa = 3.34FSKK468 pKa = 10.51FDD470 pKa = 3.37KK471 pKa = 11.04SQGRR475 pKa = 11.84LHH477 pKa = 7.01HH478 pKa = 6.99DD479 pKa = 3.25VQFRR483 pKa = 11.84LFLALGIPEE492 pKa = 4.7HH493 pKa = 6.97FVTTWFNSHH502 pKa = 5.67EE503 pKa = 4.0KK504 pKa = 10.58SYY506 pKa = 10.97IRR508 pKa = 11.84DD509 pKa = 3.57RR510 pKa = 11.84DD511 pKa = 3.73CGIGFSVDD519 pKa = 3.25YY520 pKa = 10.46QRR522 pKa = 11.84RR523 pKa = 11.84TGDD526 pKa = 2.74ACTYY530 pKa = 10.46LGNTLVTLSVLSYY543 pKa = 11.16VYY545 pKa = 10.73DD546 pKa = 4.12LSNPNILFVAASGDD560 pKa = 3.57DD561 pKa = 3.67SLIGSIEE568 pKa = 3.73PLPRR572 pKa = 11.84EE573 pKa = 4.19KK574 pKa = 10.83EE575 pKa = 4.13DD576 pKa = 3.47LCVSLFNFEE585 pKa = 4.08TKK587 pKa = 10.2FPHH590 pKa = 5.6NQPFICSKK598 pKa = 9.9FLLVVEE604 pKa = 4.98CDD606 pKa = 3.73DD607 pKa = 4.32GSEE610 pKa = 4.08EE611 pKa = 4.15VLAVPNPLKK620 pKa = 10.61LLQKK624 pKa = 10.54LGPKK628 pKa = 9.39NLQVTVLDD636 pKa = 4.97DD637 pKa = 4.31YY638 pKa = 11.44YY639 pKa = 11.6QSLCDD644 pKa = 3.84ILWVFNDD651 pKa = 3.15ADD653 pKa = 3.74VCRR656 pKa = 11.84RR657 pKa = 11.84TAEE660 pKa = 3.76LAEE663 pKa = 3.78YY664 pKa = 10.54RR665 pKa = 11.84RR666 pKa = 11.84FKK668 pKa = 9.84GTKK671 pKa = 8.33KK672 pKa = 10.63CLFLEE677 pKa = 4.46SALLSLPSLVANRR690 pKa = 11.84MKK692 pKa = 10.58FVRR695 pKa = 11.84RR696 pKa = 11.84TINLEE701 pKa = 3.62SSRR704 pKa = 11.84ACIRR708 pKa = 11.84NDD710 pKa = 3.14VYY712 pKa = 11.68SDD714 pKa = 3.93LVPHH718 pKa = 6.61FDD720 pKa = 4.21SRR722 pKa = 11.84VSRR725 pKa = 11.84CDD727 pKa = 3.36DD728 pKa = 3.32SDD730 pKa = 3.84GVRR733 pKa = 11.84TSTFDD738 pKa = 3.27DD739 pKa = 3.77RR740 pKa = 11.84KK741 pKa = 9.49SSKK744 pKa = 10.14HH745 pKa = 6.32ASDD748 pKa = 3.6KK749 pKa = 10.83LRR751 pKa = 11.84KK752 pKa = 7.13TEE754 pKa = 4.07CYY756 pKa = 10.54GEE758 pKa = 3.97ARR760 pKa = 11.84CRR762 pKa = 11.84IKK764 pKa = 10.41PRR766 pKa = 11.84RR767 pKa = 11.84NRR769 pKa = 11.84KK770 pKa = 8.99SEE772 pKa = 4.03SGAVQYY778 pKa = 10.3SQSSGIEE785 pKa = 3.72TGKK788 pKa = 10.81ANSSRR793 pKa = 11.84KK794 pKa = 9.77GRR796 pKa = 11.84IKK798 pKa = 10.7LHH800 pKa = 5.83
MM1 pKa = 7.83DD2 pKa = 5.53LVIKK6 pKa = 10.56NLIVYY11 pKa = 8.55HH12 pKa = 5.57LRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84IDD18 pKa = 2.76IGTSFGIEE26 pKa = 3.65PADD29 pKa = 3.82YY30 pKa = 10.41IDD32 pKa = 3.67WVKK35 pKa = 10.78VFLLKK40 pKa = 10.42FIIEE44 pKa = 4.12HH45 pKa = 5.26TARR48 pKa = 11.84FADD51 pKa = 4.68FATIHH56 pKa = 5.02TTMLLVLGEE65 pKa = 4.74DD66 pKa = 3.58DD67 pKa = 3.88PNYY70 pKa = 10.36VEE72 pKa = 5.19KK73 pKa = 9.84DD74 pKa = 3.61TPIMEE79 pKa = 4.98IDD81 pKa = 4.14PFYY84 pKa = 11.2LPYY87 pKa = 10.75DD88 pKa = 4.42DD89 pKa = 6.34LDD91 pKa = 3.39VDD93 pKa = 4.07YY94 pKa = 10.73TSLRR98 pKa = 11.84VCGDD102 pKa = 3.42EE103 pKa = 5.19DD104 pKa = 3.93QSCSDD109 pKa = 4.01RR110 pKa = 11.84DD111 pKa = 3.67EE112 pKa = 5.07LSDD115 pKa = 4.08FISNISHH122 pKa = 6.27IPEE125 pKa = 4.21GTSWGSEE132 pKa = 3.59SDD134 pKa = 3.47TSFVEE139 pKa = 4.1HH140 pKa = 7.24LEE142 pKa = 4.41TIQDD146 pKa = 3.34IPTKK150 pKa = 10.76CEE152 pKa = 3.63IADD155 pKa = 3.93KK156 pKa = 10.05PVEE159 pKa = 4.77EE160 pKa = 5.36IPFDD164 pKa = 4.63DD165 pKa = 5.22DD166 pKa = 4.06GKK168 pKa = 10.75VVNDD172 pKa = 3.18VWVDD176 pKa = 3.61AEE178 pKa = 4.56LSNAPEE184 pKa = 4.1ISCDD188 pKa = 3.28ADD190 pKa = 2.89IRR192 pKa = 11.84ACGFVSLRR200 pKa = 11.84LLEE203 pKa = 4.17SSRR206 pKa = 11.84GYY208 pKa = 9.84PKK210 pKa = 8.65WTPEE214 pKa = 3.93RR215 pKa = 11.84VSSGLNPDD223 pKa = 3.99LPVNSKK229 pKa = 9.96PAVDD233 pKa = 4.71EE234 pKa = 4.31IFPHH238 pKa = 5.7HH239 pKa = 6.92HH240 pKa = 6.28SVDD243 pKa = 3.3DD244 pKa = 4.14RR245 pKa = 11.84FFQEE249 pKa = 3.7WVEE252 pKa = 3.99THH254 pKa = 7.34DD255 pKa = 3.95IDD257 pKa = 6.24LEE259 pKa = 4.47VTSCDD264 pKa = 3.8LDD266 pKa = 3.23MSTFNDD272 pKa = 3.13WTKK275 pKa = 11.11GVDD278 pKa = 3.3TRR280 pKa = 11.84LVPNMSVGGLSHH292 pKa = 7.1RR293 pKa = 11.84VPTQRR298 pKa = 11.84EE299 pKa = 3.62ALLAIKK305 pKa = 9.37KK306 pKa = 10.3RR307 pKa = 11.84NMNVPEE313 pKa = 4.11LQSNFDD319 pKa = 3.87HH320 pKa = 7.95DD321 pKa = 5.07DD322 pKa = 3.38VLNRR326 pKa = 11.84CVTRR330 pKa = 11.84FITHH334 pKa = 5.6VVDD337 pKa = 3.52KK338 pKa = 10.93TRR340 pKa = 11.84LSKK343 pKa = 10.94LNPISGEE350 pKa = 3.79EE351 pKa = 3.48LHH353 pKa = 6.79YY354 pKa = 10.77FNQYY358 pKa = 10.65LEE360 pKa = 4.47NKK362 pKa = 9.44NPPLSEE368 pKa = 4.1YY369 pKa = 10.16KK370 pKa = 10.95GPVPLVALDD379 pKa = 4.49KK380 pKa = 10.26YY381 pKa = 9.66MHH383 pKa = 6.57MIKK386 pKa = 7.75TTLKK390 pKa = 10.38PVEE393 pKa = 4.37EE394 pKa = 4.96DD395 pKa = 3.04NLHH398 pKa = 6.21IEE400 pKa = 4.23RR401 pKa = 11.84PIPATITYY409 pKa = 8.34HH410 pKa = 5.89KK411 pKa = 9.69KK412 pKa = 9.51GVVMMTSPYY421 pKa = 9.61FLCAMVRR428 pKa = 11.84LLYY431 pKa = 10.08VLKK434 pKa = 10.74SKK436 pKa = 10.49FVVPTGKK443 pKa = 9.74YY444 pKa = 9.12HH445 pKa = 7.59QIFQMNPEE453 pKa = 4.33LLKK456 pKa = 10.62HH457 pKa = 5.58SKK459 pKa = 8.92EE460 pKa = 3.97FKK462 pKa = 10.52EE463 pKa = 3.83IDD465 pKa = 3.34FSKK468 pKa = 10.51FDD470 pKa = 3.37KK471 pKa = 11.04SQGRR475 pKa = 11.84LHH477 pKa = 7.01HH478 pKa = 6.99DD479 pKa = 3.25VQFRR483 pKa = 11.84LFLALGIPEE492 pKa = 4.7HH493 pKa = 6.97FVTTWFNSHH502 pKa = 5.67EE503 pKa = 4.0KK504 pKa = 10.58SYY506 pKa = 10.97IRR508 pKa = 11.84DD509 pKa = 3.57RR510 pKa = 11.84DD511 pKa = 3.73CGIGFSVDD519 pKa = 3.25YY520 pKa = 10.46QRR522 pKa = 11.84RR523 pKa = 11.84TGDD526 pKa = 2.74ACTYY530 pKa = 10.46LGNTLVTLSVLSYY543 pKa = 11.16VYY545 pKa = 10.73DD546 pKa = 4.12LSNPNILFVAASGDD560 pKa = 3.57DD561 pKa = 3.67SLIGSIEE568 pKa = 3.73PLPRR572 pKa = 11.84EE573 pKa = 4.19KK574 pKa = 10.83EE575 pKa = 4.13DD576 pKa = 3.47LCVSLFNFEE585 pKa = 4.08TKK587 pKa = 10.2FPHH590 pKa = 5.6NQPFICSKK598 pKa = 9.9FLLVVEE604 pKa = 4.98CDD606 pKa = 3.73DD607 pKa = 4.32GSEE610 pKa = 4.08EE611 pKa = 4.15VLAVPNPLKK620 pKa = 10.61LLQKK624 pKa = 10.54LGPKK628 pKa = 9.39NLQVTVLDD636 pKa = 4.97DD637 pKa = 4.31YY638 pKa = 11.44YY639 pKa = 11.6QSLCDD644 pKa = 3.84ILWVFNDD651 pKa = 3.15ADD653 pKa = 3.74VCRR656 pKa = 11.84RR657 pKa = 11.84TAEE660 pKa = 3.76LAEE663 pKa = 3.78YY664 pKa = 10.54RR665 pKa = 11.84RR666 pKa = 11.84FKK668 pKa = 9.84GTKK671 pKa = 8.33KK672 pKa = 10.63CLFLEE677 pKa = 4.46SALLSLPSLVANRR690 pKa = 11.84MKK692 pKa = 10.58FVRR695 pKa = 11.84RR696 pKa = 11.84TINLEE701 pKa = 3.62SSRR704 pKa = 11.84ACIRR708 pKa = 11.84NDD710 pKa = 3.14VYY712 pKa = 11.68SDD714 pKa = 3.93LVPHH718 pKa = 6.61FDD720 pKa = 4.21SRR722 pKa = 11.84VSRR725 pKa = 11.84CDD727 pKa = 3.36DD728 pKa = 3.32SDD730 pKa = 3.84GVRR733 pKa = 11.84TSTFDD738 pKa = 3.27DD739 pKa = 3.77RR740 pKa = 11.84KK741 pKa = 9.49SSKK744 pKa = 10.14HH745 pKa = 6.32ASDD748 pKa = 3.6KK749 pKa = 10.83LRR751 pKa = 11.84KK752 pKa = 7.13TEE754 pKa = 4.07CYY756 pKa = 10.54GEE758 pKa = 3.97ARR760 pKa = 11.84CRR762 pKa = 11.84IKK764 pKa = 10.41PRR766 pKa = 11.84RR767 pKa = 11.84NRR769 pKa = 11.84KK770 pKa = 8.99SEE772 pKa = 4.03SGAVQYY778 pKa = 10.3SQSSGIEE785 pKa = 3.72TGKK788 pKa = 10.81ANSSRR793 pKa = 11.84KK794 pKa = 9.77GRR796 pKa = 11.84IKK798 pKa = 10.7LHH800 pKa = 5.83
Molecular weight: 91.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P89678|RDRP_TOBSV RNA-directed RNA polymerase 2a OS=Tobacco streak virus (strain WC) OX=12318 GN=ORF2a PE=3 SV=1
MM1 pKa = 6.93NTLIQGPDD9 pKa = 3.26HH10 pKa = 7.37PSNAMSSRR18 pKa = 11.84ANNRR22 pKa = 11.84SNNSRR27 pKa = 11.84CPTCIDD33 pKa = 4.0EE34 pKa = 4.81LDD36 pKa = 3.45AMARR40 pKa = 11.84NCPAHH45 pKa = 5.55NTVNTVSRR53 pKa = 11.84RR54 pKa = 11.84QRR56 pKa = 11.84RR57 pKa = 11.84NAARR61 pKa = 11.84AAAYY65 pKa = 9.79RR66 pKa = 11.84NANARR71 pKa = 11.84VPLPLPVVSVSRR83 pKa = 11.84PQAKK87 pKa = 10.2ASLRR91 pKa = 11.84LPNNQVWVTRR101 pKa = 11.84KK102 pKa = 9.79ASEE105 pKa = 3.9WSAKK109 pKa = 9.28TVDD112 pKa = 3.85TNDD115 pKa = 4.54AIPFKK120 pKa = 10.15TIVEE124 pKa = 4.91GIPEE128 pKa = 4.17IGAEE132 pKa = 4.19TKK134 pKa = 10.45FFRR137 pKa = 11.84LLIGFVAVSDD147 pKa = 4.06GTFGMVDD154 pKa = 3.6GVTGDD159 pKa = 4.29VIPDD163 pKa = 3.61PPVVGRR169 pKa = 11.84LGFKK173 pKa = 10.51KK174 pKa = 9.03NTYY177 pKa = 9.52RR178 pKa = 11.84SRR180 pKa = 11.84DD181 pKa = 3.17FDD183 pKa = 4.71LGGKK187 pKa = 9.19LLNQLDD193 pKa = 3.73DD194 pKa = 4.66RR195 pKa = 11.84AVVWCLDD202 pKa = 3.29EE203 pKa = 4.46RR204 pKa = 11.84RR205 pKa = 11.84RR206 pKa = 11.84EE207 pKa = 4.1AKK209 pKa = 9.57RR210 pKa = 11.84VQLAGYY216 pKa = 8.3WIAISKK222 pKa = 7.65PAPLMPPEE230 pKa = 5.11DD231 pKa = 3.6FLVNQDD237 pKa = 3.07
MM1 pKa = 6.93NTLIQGPDD9 pKa = 3.26HH10 pKa = 7.37PSNAMSSRR18 pKa = 11.84ANNRR22 pKa = 11.84SNNSRR27 pKa = 11.84CPTCIDD33 pKa = 4.0EE34 pKa = 4.81LDD36 pKa = 3.45AMARR40 pKa = 11.84NCPAHH45 pKa = 5.55NTVNTVSRR53 pKa = 11.84RR54 pKa = 11.84QRR56 pKa = 11.84RR57 pKa = 11.84NAARR61 pKa = 11.84AAAYY65 pKa = 9.79RR66 pKa = 11.84NANARR71 pKa = 11.84VPLPLPVVSVSRR83 pKa = 11.84PQAKK87 pKa = 10.2ASLRR91 pKa = 11.84LPNNQVWVTRR101 pKa = 11.84KK102 pKa = 9.79ASEE105 pKa = 3.9WSAKK109 pKa = 9.28TVDD112 pKa = 3.85TNDD115 pKa = 4.54AIPFKK120 pKa = 10.15TIVEE124 pKa = 4.91GIPEE128 pKa = 4.17IGAEE132 pKa = 4.19TKK134 pKa = 10.45FFRR137 pKa = 11.84LLIGFVAVSDD147 pKa = 4.06GTFGMVDD154 pKa = 3.6GVTGDD159 pKa = 4.29VIPDD163 pKa = 3.61PPVVGRR169 pKa = 11.84LGFKK173 pKa = 10.51KK174 pKa = 9.03NTYY177 pKa = 9.52RR178 pKa = 11.84SRR180 pKa = 11.84DD181 pKa = 3.17FDD183 pKa = 4.71LGGKK187 pKa = 9.19LLNQLDD193 pKa = 3.73DD194 pKa = 4.66RR195 pKa = 11.84AVVWCLDD202 pKa = 3.29EE203 pKa = 4.46RR204 pKa = 11.84RR205 pKa = 11.84RR206 pKa = 11.84EE207 pKa = 4.1AKK209 pKa = 9.57RR210 pKa = 11.84VQLAGYY216 pKa = 8.3WIAISKK222 pKa = 7.65PAPLMPPEE230 pKa = 5.11DD231 pKa = 3.6FLVNQDD237 pKa = 3.07
Molecular weight: 26.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2625 |
205 |
1094 |
525.0 |
59.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.248 ± 1.081 | 2.286 ± 0.264 |
6.933 ± 0.839 | 6.095 ± 0.351 |
4.305 ± 0.334 | 4.533 ± 0.282 |
2.743 ± 0.46 | 5.905 ± 0.388 |
6.248 ± 0.345 | 9.105 ± 0.276 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.514 ± 0.368 | 4.305 ± 0.442 |
5.181 ± 0.777 | 2.324 ± 0.129 |
5.79 ± 0.683 | 8.114 ± 0.366 |
5.79 ± 0.644 | 8.038 ± 0.298 |
0.952 ± 0.112 | 2.59 ± 0.518 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
