
Fowl adenovirus
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Rowavirales; Adenoviridae; Aviadenovirus; unclassified Aviadenovirus
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
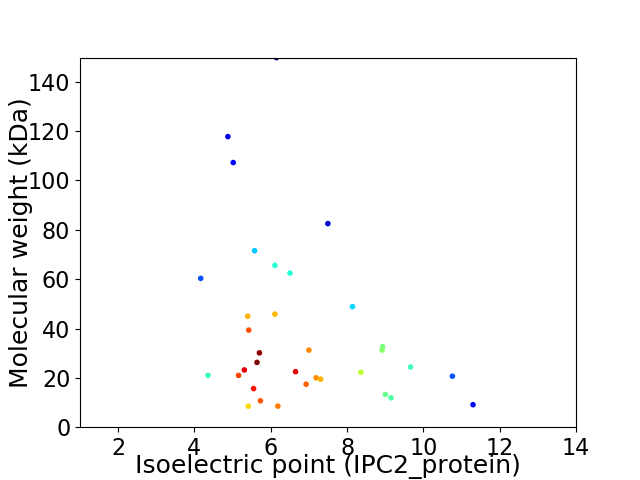
Virtual 2D-PAGE plot for 34 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5P8NQN0|A0A5P8NQN0_9ADEN 22k protein OS=Fowl adenovirus OX=1354736 PE=4 SV=1
MM1 pKa = 7.52AKK3 pKa = 8.76STPFTFSMGQHH14 pKa = 6.21SSRR17 pKa = 11.84KK18 pKa = 9.39RR19 pKa = 11.84PADD22 pKa = 3.44SEE24 pKa = 4.19NTQNASKK31 pKa = 10.45VAKK34 pKa = 8.38TQTSATRR41 pKa = 11.84AGVDD45 pKa = 3.89GNDD48 pKa = 4.54DD49 pKa = 4.1LNLVYY54 pKa = 9.94PFWLQNSTSGGGGGGSGGNPSLNPPFIDD82 pKa = 3.73PNGPLYY88 pKa = 10.31VQNSLLYY95 pKa = 10.35VKK97 pKa = 7.76TTAPIEE103 pKa = 4.16VEE105 pKa = 4.18NKK107 pKa = 10.27SLALAYY113 pKa = 10.4NSSLDD118 pKa = 3.5VDD120 pKa = 4.2AQNQLQVKK128 pKa = 9.2VDD130 pKa = 3.45AEE132 pKa = 4.46GPIRR136 pKa = 11.84ISPDD140 pKa = 3.08GLDD143 pKa = 3.18IAVDD147 pKa = 4.11PSTLEE152 pKa = 4.49VDD154 pKa = 4.89DD155 pKa = 4.35EE156 pKa = 4.52WEE158 pKa = 4.23LTVKK162 pKa = 10.5LDD164 pKa = 3.59PAGPISSSSAGINIRR179 pKa = 11.84VDD181 pKa = 3.33DD182 pKa = 4.2TLLIEE187 pKa = 6.18DD188 pKa = 5.61DD189 pKa = 4.27DD190 pKa = 4.23TTQVKK195 pKa = 9.92EE196 pKa = 4.4LGVHH200 pKa = 6.68LNPNGPITADD210 pKa = 3.16QDD212 pKa = 4.14GLDD215 pKa = 4.68LEE217 pKa = 5.14VDD219 pKa = 3.83PQTLTVTTSGATGGVLGVLLKK240 pKa = 10.67PSGGLQTSIQGIGVAVADD258 pKa = 4.18TLTISSNNTVEE269 pKa = 4.99VKK271 pKa = 9.88TDD273 pKa = 3.71PNGSIGSSSNGIAVVTDD290 pKa = 3.45PAGPLTTSSNGLSLKK305 pKa = 8.58LTPNGSIQSSSTGLSVQTDD324 pKa = 3.58PAGPITSGANGLSLSYY340 pKa = 9.5DD341 pKa = 3.29TSDD344 pKa = 3.48FTVSQGMLSIIRR356 pKa = 11.84NPSTYY361 pKa = 9.77PDD363 pKa = 3.91AYY365 pKa = 10.87LEE367 pKa = 4.61SGTNLLNNYY376 pKa = 7.05TAYY379 pKa = 10.66AEE381 pKa = 4.05NSSNYY386 pKa = 9.63KK387 pKa = 9.97FNCAYY392 pKa = 10.3FLQSWYY398 pKa = 11.27SNGLVTSSLYY408 pKa = 11.07LKK410 pKa = 10.39INRR413 pKa = 11.84DD414 pKa = 3.68NLTSLPSGQLSEE426 pKa = 3.98NAKK429 pKa = 10.55YY430 pKa = 8.24FTFWVPTYY438 pKa = 11.17EE439 pKa = 5.04SMNLSNVATPTITPSSVPWGAFLPAQNCTSNPAFKK474 pKa = 10.69YY475 pKa = 10.75YY476 pKa = 9.51LTQPPSIYY484 pKa = 10.32FEE486 pKa = 4.53PEE488 pKa = 3.66SGSVQTFQPVLTGDD502 pKa = 3.49WDD504 pKa = 4.24TNTYY508 pKa = 10.85NPGTVQVCILPQTVVGGQSTFVNMTCYY535 pKa = 10.52NFRR538 pKa = 11.84CQNPGIFKK546 pKa = 10.24VAASSGTFTIGPIFYY561 pKa = 10.31SCPTNKK567 pKa = 8.58LTQPP571 pKa = 3.58
MM1 pKa = 7.52AKK3 pKa = 8.76STPFTFSMGQHH14 pKa = 6.21SSRR17 pKa = 11.84KK18 pKa = 9.39RR19 pKa = 11.84PADD22 pKa = 3.44SEE24 pKa = 4.19NTQNASKK31 pKa = 10.45VAKK34 pKa = 8.38TQTSATRR41 pKa = 11.84AGVDD45 pKa = 3.89GNDD48 pKa = 4.54DD49 pKa = 4.1LNLVYY54 pKa = 9.94PFWLQNSTSGGGGGGSGGNPSLNPPFIDD82 pKa = 3.73PNGPLYY88 pKa = 10.31VQNSLLYY95 pKa = 10.35VKK97 pKa = 7.76TTAPIEE103 pKa = 4.16VEE105 pKa = 4.18NKK107 pKa = 10.27SLALAYY113 pKa = 10.4NSSLDD118 pKa = 3.5VDD120 pKa = 4.2AQNQLQVKK128 pKa = 9.2VDD130 pKa = 3.45AEE132 pKa = 4.46GPIRR136 pKa = 11.84ISPDD140 pKa = 3.08GLDD143 pKa = 3.18IAVDD147 pKa = 4.11PSTLEE152 pKa = 4.49VDD154 pKa = 4.89DD155 pKa = 4.35EE156 pKa = 4.52WEE158 pKa = 4.23LTVKK162 pKa = 10.5LDD164 pKa = 3.59PAGPISSSSAGINIRR179 pKa = 11.84VDD181 pKa = 3.33DD182 pKa = 4.2TLLIEE187 pKa = 6.18DD188 pKa = 5.61DD189 pKa = 4.27DD190 pKa = 4.23TTQVKK195 pKa = 9.92EE196 pKa = 4.4LGVHH200 pKa = 6.68LNPNGPITADD210 pKa = 3.16QDD212 pKa = 4.14GLDD215 pKa = 4.68LEE217 pKa = 5.14VDD219 pKa = 3.83PQTLTVTTSGATGGVLGVLLKK240 pKa = 10.67PSGGLQTSIQGIGVAVADD258 pKa = 4.18TLTISSNNTVEE269 pKa = 4.99VKK271 pKa = 9.88TDD273 pKa = 3.71PNGSIGSSSNGIAVVTDD290 pKa = 3.45PAGPLTTSSNGLSLKK305 pKa = 8.58LTPNGSIQSSSTGLSVQTDD324 pKa = 3.58PAGPITSGANGLSLSYY340 pKa = 9.5DD341 pKa = 3.29TSDD344 pKa = 3.48FTVSQGMLSIIRR356 pKa = 11.84NPSTYY361 pKa = 9.77PDD363 pKa = 3.91AYY365 pKa = 10.87LEE367 pKa = 4.61SGTNLLNNYY376 pKa = 7.05TAYY379 pKa = 10.66AEE381 pKa = 4.05NSSNYY386 pKa = 9.63KK387 pKa = 9.97FNCAYY392 pKa = 10.3FLQSWYY398 pKa = 11.27SNGLVTSSLYY408 pKa = 11.07LKK410 pKa = 10.39INRR413 pKa = 11.84DD414 pKa = 3.68NLTSLPSGQLSEE426 pKa = 3.98NAKK429 pKa = 10.55YY430 pKa = 8.24FTFWVPTYY438 pKa = 11.17EE439 pKa = 5.04SMNLSNVATPTITPSSVPWGAFLPAQNCTSNPAFKK474 pKa = 10.69YY475 pKa = 10.75YY476 pKa = 9.51LTQPPSIYY484 pKa = 10.32FEE486 pKa = 4.53PEE488 pKa = 3.66SGSVQTFQPVLTGDD502 pKa = 3.49WDD504 pKa = 4.24TNTYY508 pKa = 10.85NPGTVQVCILPQTVVGGQSTFVNMTCYY535 pKa = 10.52NFRR538 pKa = 11.84CQNPGIFKK546 pKa = 10.24VAASSGTFTIGPIFYY561 pKa = 10.31SCPTNKK567 pKa = 8.58LTQPP571 pKa = 3.58
Molecular weight: 60.33 kDa
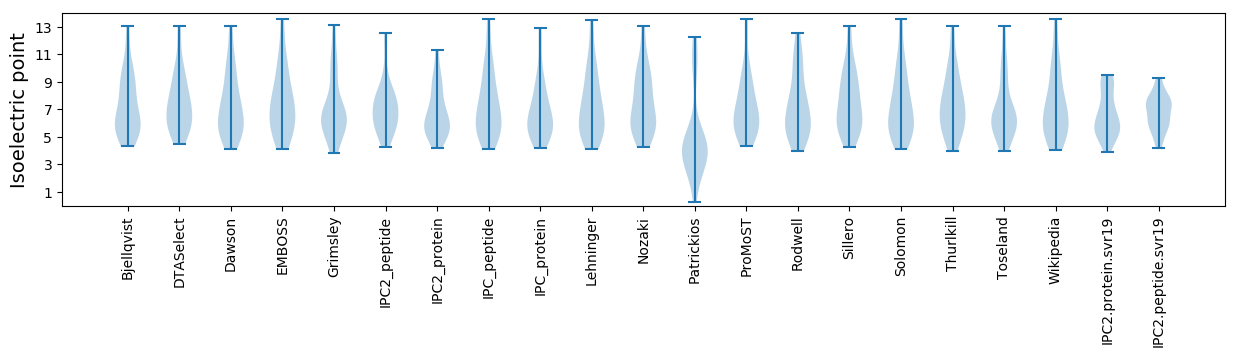
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5P8NQL4|A0A5P8NQL4_9ADEN PTP protein OS=Fowl adenovirus OX=1354736 PE=4 SV=1
MM1 pKa = 7.68SILISPNDD9 pKa = 3.25NRR11 pKa = 11.84GWGMRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84SRR20 pKa = 11.84SSMRR24 pKa = 11.84GVGIRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84LTLRR36 pKa = 11.84TLLGLGTSSRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84SGGRR52 pKa = 11.84SSRR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 11.84SRR59 pKa = 11.84PASTTTRR66 pKa = 11.84VMLVRR71 pKa = 11.84TSRR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 3.32
MM1 pKa = 7.68SILISPNDD9 pKa = 3.25NRR11 pKa = 11.84GWGMRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84SRR20 pKa = 11.84SSMRR24 pKa = 11.84GVGIRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84LTLRR36 pKa = 11.84TLLGLGTSSRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84SGGRR52 pKa = 11.84SSRR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 11.84SRR59 pKa = 11.84PASTTTRR66 pKa = 11.84VMLVRR71 pKa = 11.84TSRR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 3.32
Molecular weight: 9.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
11899 |
76 |
1305 |
350.0 |
39.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.841 ± 0.442 | 1.79 ± 0.26 |
5.572 ± 0.249 | 5.95 ± 0.52 |
4.152 ± 0.269 | 6.429 ± 0.316 |
2.378 ± 0.278 | 3.958 ± 0.174 |
3.706 ± 0.314 | 8.673 ± 0.334 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.395 ± 0.187 | 4.379 ± 0.487 |
6.454 ± 0.246 | 3.664 ± 0.239 |
7.37 ± 0.54 | 7.127 ± 0.424 |
6.043 ± 0.383 | 6.656 ± 0.277 |
1.361 ± 0.109 | 4.101 ± 0.285 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
