
Leptospira phage LE4
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Nylescharonvirus; Leptospira virus LE4
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
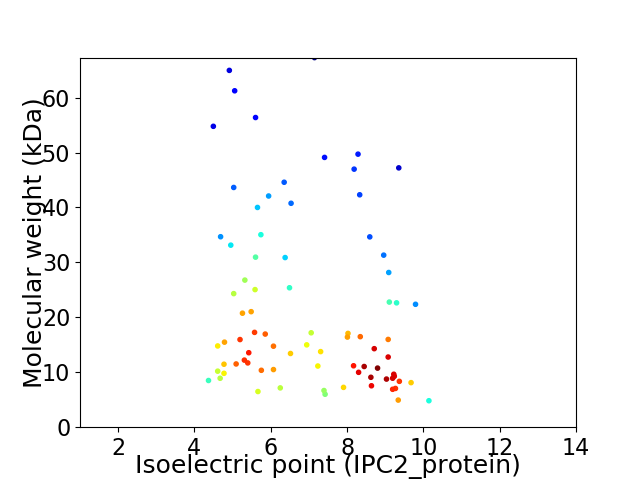
Virtual 2D-PAGE plot for 81 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A343LEE5|A0A343LEE5_9CAUD Uncharacterized protein OS=Leptospira phage LE4 OX=2041383 PE=4 SV=1
MM1 pKa = 7.44SNKK4 pKa = 9.72KK5 pKa = 9.94VIQYY9 pKa = 7.12STKK12 pKa = 11.02DD13 pKa = 3.57FLTALADD20 pKa = 3.58IEE22 pKa = 4.6SDD24 pKa = 3.41PEE26 pKa = 4.11LRR28 pKa = 11.84KK29 pKa = 9.9FPARR33 pKa = 11.84FKK35 pKa = 10.35RR36 pKa = 11.84LHH38 pKa = 6.67AGTVDD43 pKa = 4.99AINNNLNAVFNSIILRR59 pKa = 11.84TAFSRR64 pKa = 11.84PILQDD69 pKa = 2.88ILQLIDD75 pKa = 4.08YY76 pKa = 9.44QMSWKK81 pKa = 7.96KK82 pKa = 9.22TSVATIQITVVSSATPYY99 pKa = 10.77VILKK103 pKa = 9.68ADD105 pKa = 4.68CIFGTAGTVDD115 pKa = 3.19QASIQFEE122 pKa = 4.29SRR124 pKa = 11.84EE125 pKa = 4.33DD126 pKa = 3.34IAVPALTTLISRR138 pKa = 11.84TVYY141 pKa = 10.7SQTTQPQRR149 pKa = 11.84TIGVTDD155 pKa = 4.17GSNFQEE161 pKa = 4.47IDD163 pKa = 4.05LPDD166 pKa = 5.32LDD168 pKa = 4.9ILPEE172 pKa = 4.08TLFITINGNQYY183 pKa = 11.53DD184 pKa = 4.24LVDD187 pKa = 4.09SFAEE191 pKa = 4.11SASTDD196 pKa = 2.89RR197 pKa = 11.84HH198 pKa = 4.06FRR200 pKa = 11.84IYY202 pKa = 10.36FRR204 pKa = 11.84SDD206 pKa = 2.53GSSYY210 pKa = 10.52IQLPGVDD217 pKa = 3.55QVDD220 pKa = 3.62NTSFGQVPPQGQTVYY235 pKa = 11.36ANYY238 pKa = 8.89ATGGGANTNVSANTITEE255 pKa = 4.07YY256 pKa = 11.1LGTDD260 pKa = 3.5SNIVSVNNGSPASGGLDD277 pKa = 3.08EE278 pKa = 5.06EE279 pKa = 5.1TIQNAVTIAPLRR291 pKa = 11.84ARR293 pKa = 11.84TTRR296 pKa = 11.84YY297 pKa = 9.4FINRR301 pKa = 11.84STGVAVAKK309 pKa = 10.46EE310 pKa = 3.96IDD312 pKa = 3.48GVLYY316 pKa = 10.93ADD318 pKa = 3.58IVKK321 pKa = 10.55VGALSCEE328 pKa = 4.68CYY330 pKa = 10.17IIPVGGGIASSQLLQEE346 pKa = 4.42VEE348 pKa = 4.28DD349 pKa = 4.36LLVSRR354 pKa = 11.84SPLEE358 pKa = 4.23EE359 pKa = 4.25IEE361 pKa = 4.39VNAYY365 pKa = 5.93TASYY369 pKa = 8.49LTTFVNMQIKK379 pKa = 9.97LLPGFSYY386 pKa = 11.57ADD388 pKa = 3.32VAKK391 pKa = 10.29FSSLAVCQTLHH402 pKa = 6.86VLGQYY407 pKa = 9.91IFEE410 pKa = 4.49VYY412 pKa = 10.48ASNGVQSAIEE422 pKa = 4.65EE423 pKa = 4.29INSIYY428 pKa = 10.78GPIIGYY434 pKa = 7.21TFTIADD440 pKa = 3.69SSGIIQILGNVEE452 pKa = 3.85TAVVSKK458 pKa = 9.84PRR460 pKa = 11.84KK461 pKa = 8.39ISEE464 pKa = 3.77IYY466 pKa = 8.3MALGFVQGLDD476 pKa = 3.45YY477 pKa = 11.62ANIVAPNSDD486 pKa = 3.74LVPNNGSITNVSTVTLTQVNN506 pKa = 3.76
MM1 pKa = 7.44SNKK4 pKa = 9.72KK5 pKa = 9.94VIQYY9 pKa = 7.12STKK12 pKa = 11.02DD13 pKa = 3.57FLTALADD20 pKa = 3.58IEE22 pKa = 4.6SDD24 pKa = 3.41PEE26 pKa = 4.11LRR28 pKa = 11.84KK29 pKa = 9.9FPARR33 pKa = 11.84FKK35 pKa = 10.35RR36 pKa = 11.84LHH38 pKa = 6.67AGTVDD43 pKa = 4.99AINNNLNAVFNSIILRR59 pKa = 11.84TAFSRR64 pKa = 11.84PILQDD69 pKa = 2.88ILQLIDD75 pKa = 4.08YY76 pKa = 9.44QMSWKK81 pKa = 7.96KK82 pKa = 9.22TSVATIQITVVSSATPYY99 pKa = 10.77VILKK103 pKa = 9.68ADD105 pKa = 4.68CIFGTAGTVDD115 pKa = 3.19QASIQFEE122 pKa = 4.29SRR124 pKa = 11.84EE125 pKa = 4.33DD126 pKa = 3.34IAVPALTTLISRR138 pKa = 11.84TVYY141 pKa = 10.7SQTTQPQRR149 pKa = 11.84TIGVTDD155 pKa = 4.17GSNFQEE161 pKa = 4.47IDD163 pKa = 4.05LPDD166 pKa = 5.32LDD168 pKa = 4.9ILPEE172 pKa = 4.08TLFITINGNQYY183 pKa = 11.53DD184 pKa = 4.24LVDD187 pKa = 4.09SFAEE191 pKa = 4.11SASTDD196 pKa = 2.89RR197 pKa = 11.84HH198 pKa = 4.06FRR200 pKa = 11.84IYY202 pKa = 10.36FRR204 pKa = 11.84SDD206 pKa = 2.53GSSYY210 pKa = 10.52IQLPGVDD217 pKa = 3.55QVDD220 pKa = 3.62NTSFGQVPPQGQTVYY235 pKa = 11.36ANYY238 pKa = 8.89ATGGGANTNVSANTITEE255 pKa = 4.07YY256 pKa = 11.1LGTDD260 pKa = 3.5SNIVSVNNGSPASGGLDD277 pKa = 3.08EE278 pKa = 5.06EE279 pKa = 5.1TIQNAVTIAPLRR291 pKa = 11.84ARR293 pKa = 11.84TTRR296 pKa = 11.84YY297 pKa = 9.4FINRR301 pKa = 11.84STGVAVAKK309 pKa = 10.46EE310 pKa = 3.96IDD312 pKa = 3.48GVLYY316 pKa = 10.93ADD318 pKa = 3.58IVKK321 pKa = 10.55VGALSCEE328 pKa = 4.68CYY330 pKa = 10.17IIPVGGGIASSQLLQEE346 pKa = 4.42VEE348 pKa = 4.28DD349 pKa = 4.36LLVSRR354 pKa = 11.84SPLEE358 pKa = 4.23EE359 pKa = 4.25IEE361 pKa = 4.39VNAYY365 pKa = 5.93TASYY369 pKa = 8.49LTTFVNMQIKK379 pKa = 9.97LLPGFSYY386 pKa = 11.57ADD388 pKa = 3.32VAKK391 pKa = 10.29FSSLAVCQTLHH402 pKa = 6.86VLGQYY407 pKa = 9.91IFEE410 pKa = 4.49VYY412 pKa = 10.48ASNGVQSAIEE422 pKa = 4.65EE423 pKa = 4.29INSIYY428 pKa = 10.78GPIIGYY434 pKa = 7.21TFTIADD440 pKa = 3.69SSGIIQILGNVEE452 pKa = 3.85TAVVSKK458 pKa = 9.84PRR460 pKa = 11.84KK461 pKa = 8.39ISEE464 pKa = 3.77IYY466 pKa = 8.3MALGFVQGLDD476 pKa = 3.45YY477 pKa = 11.62ANIVAPNSDD486 pKa = 3.74LVPNNGSITNVSTVTLTQVNN506 pKa = 3.76
Molecular weight: 54.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A343LEH1|A0A343LEH1_9CAUD Uncharacterized protein OS=Leptospira phage LE4 OX=2041383 PE=4 SV=1
MM1 pKa = 8.45IEE3 pKa = 3.75MPGYY7 pKa = 10.3IFILMMMIFWSLGYY21 pKa = 11.29LMGKK25 pKa = 9.82RR26 pKa = 11.84SAKK29 pKa = 9.57KK30 pKa = 10.12RR31 pKa = 11.84PAVHH35 pKa = 6.27NKK37 pKa = 9.92SSVRR41 pKa = 3.54
MM1 pKa = 8.45IEE3 pKa = 3.75MPGYY7 pKa = 10.3IFILMMMIFWSLGYY21 pKa = 11.29LMGKK25 pKa = 9.82RR26 pKa = 11.84SAKK29 pKa = 9.57KK30 pKa = 10.12RR31 pKa = 11.84PAVHH35 pKa = 6.27NKK37 pKa = 9.92SSVRR41 pKa = 3.54
Molecular weight: 4.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
15527 |
41 |
592 |
191.7 |
21.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.255 ± 0.285 | 0.915 ± 0.116 |
5.326 ± 0.216 | 8.141 ± 0.507 |
4.772 ± 0.229 | 6.157 ± 0.423 |
1.591 ± 0.152 | 7.548 ± 0.301 |
9.719 ± 0.573 | 7.703 ± 0.194 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.312 ± 0.191 | 5.32 ± 0.18 |
3.703 ± 0.176 | 4.115 ± 0.185 |
4.103 ± 0.149 | 6.994 ± 0.275 |
5.706 ± 0.346 | 5.874 ± 0.263 |
1.069 ± 0.093 | 3.677 ± 0.162 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
