
Desulfovibrio sp. OH1186_COT-070
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfovibrionales; Desulfovibrionaceae; Desulfovibrio; unclassified Desulfovibrio
Average proteome isoelectric point is 6.9
Get precalculated fractions of proteins
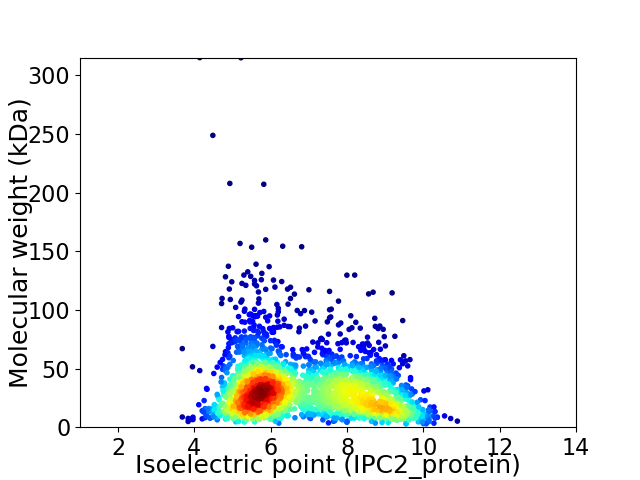
Virtual 2D-PAGE plot for 2373 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3P2A4F8|A0A3P2A4F8_9DELT Uncharacterized protein OS=Desulfovibrio sp. OH1186_COT-070 OX=2491045 GN=EII23_00570 PE=4 SV=1
MM1 pKa = 7.12QNKK4 pKa = 9.61EE5 pKa = 4.13YY6 pKa = 10.9LHH8 pKa = 6.66AAAKK12 pKa = 10.47AIAQQIEE19 pKa = 4.4GNLLVIGVSLDD30 pKa = 3.56PDD32 pKa = 3.45VADD35 pKa = 3.29YY36 pKa = 10.32MGAFEE41 pKa = 5.04EE42 pKa = 4.49EE43 pKa = 4.98AITLADD49 pKa = 3.69VVEE52 pKa = 5.26DD53 pKa = 3.94GLLTVTNSGEE63 pKa = 3.92VMYY66 pKa = 10.99VGNNN70 pKa = 3.11
MM1 pKa = 7.12QNKK4 pKa = 9.61EE5 pKa = 4.13YY6 pKa = 10.9LHH8 pKa = 6.66AAAKK12 pKa = 10.47AIAQQIEE19 pKa = 4.4GNLLVIGVSLDD30 pKa = 3.56PDD32 pKa = 3.45VADD35 pKa = 3.29YY36 pKa = 10.32MGAFEE41 pKa = 5.04EE42 pKa = 4.49EE43 pKa = 4.98AITLADD49 pKa = 3.69VVEE52 pKa = 5.26DD53 pKa = 3.94GLLTVTNSGEE63 pKa = 3.92VMYY66 pKa = 10.99VGNNN70 pKa = 3.11
Molecular weight: 7.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3P1ZXK6|A0A3P1ZXK6_9DELT Recombinase family protein OS=Desulfovibrio sp. OH1186_COT-070 OX=2491045 GN=EII23_03890 PE=4 SV=1
MM1 pKa = 7.76RR2 pKa = 11.84HH3 pKa = 6.22PPLPAPLLWQSLLGFWILGILAVAWPVPSASCAFLLFLADD43 pKa = 3.36ARR45 pKa = 11.84LWRR48 pKa = 11.84AKK50 pKa = 10.77GIFAALAVLLAGVLAGHH67 pKa = 7.22LTTGKK72 pKa = 8.18MWRR75 pKa = 11.84DD76 pKa = 3.44HH77 pKa = 6.32TPTPVAWLDD86 pKa = 3.62TAPKK90 pKa = 10.3SMRR93 pKa = 11.84ICGQIQEE100 pKa = 4.39VRR102 pKa = 11.84GLPDD106 pKa = 2.93NRR108 pKa = 11.84IRR110 pKa = 11.84MLLADD115 pKa = 4.02VRR117 pKa = 11.84PHH119 pKa = 6.47GGAEE123 pKa = 3.96KK124 pKa = 10.52SLSGLMEE131 pKa = 4.11WTWQNPSAYY140 pKa = 8.68PLPGQSACLSRR151 pKa = 11.84RR152 pKa = 11.84PQAMQGFANPGLPDD166 pKa = 4.01RR167 pKa = 11.84DD168 pKa = 3.52VVIQSVRR175 pKa = 11.84NVRR178 pKa = 11.84WRR180 pKa = 11.84IWSQGEE186 pKa = 4.12SGQPRR191 pKa = 11.84IDD193 pKa = 3.75GQPSLSARR201 pKa = 11.84WRR203 pKa = 11.84EE204 pKa = 3.97SLRR207 pKa = 11.84QGFFTVLSPDD217 pKa = 3.5AEE219 pKa = 4.16HH220 pKa = 7.26FPRR223 pKa = 11.84QKK225 pKa = 10.64LPQGKK230 pKa = 9.82AILVALLFGDD240 pKa = 4.4RR241 pKa = 11.84QFLDD245 pKa = 3.36NQTIAGFSMAALGHH259 pKa = 5.87SLALSGQHH267 pKa = 6.35LAVAGLAGILLVVVVARR284 pKa = 11.84AYY286 pKa = 9.86PGIYY290 pKa = 9.79LCRR293 pKa = 11.84TRR295 pKa = 11.84TVWVMSASVPPALAYY310 pKa = 10.37LWLGNAPPSLVRR322 pKa = 11.84ATIMLCILALWLLRR336 pKa = 11.84RR337 pKa = 11.84RR338 pKa = 11.84VHH340 pKa = 5.12TTLDD344 pKa = 3.73VLCSALFCITLFSPAAVFDD363 pKa = 3.81TGLQLSAICVAVIGMSMPWLRR384 pKa = 11.84RR385 pKa = 11.84LLPRR389 pKa = 11.84PHH391 pKa = 6.91PAEE394 pKa = 3.99HH395 pKa = 6.4AARR398 pKa = 11.84WRR400 pKa = 11.84SLLQDD405 pKa = 4.43ALWSLQRR412 pKa = 11.84IFALSLIIQTVLLPVGILLFGNTGHH437 pKa = 7.51WFPLNVLWLPLVNLLVLPGAALGLLAGACGWDD469 pKa = 3.18AAARR473 pKa = 11.84AILEE477 pKa = 4.34YY478 pKa = 10.93ASLPCQWLADD488 pKa = 3.83ALAWLARR495 pKa = 11.84QNALEE500 pKa = 4.23SPVVLRR506 pKa = 11.84PHH508 pKa = 6.04WTALPAFAALTAAAALRR525 pKa = 11.84GGRR528 pKa = 11.84LRR530 pKa = 11.84LPPAGRR536 pKa = 11.84RR537 pKa = 11.84LLLAGALLLCAGPLLRR553 pKa = 11.84AGEE556 pKa = 4.13RR557 pKa = 11.84LSPEE561 pKa = 3.61VRR563 pKa = 11.84LDD565 pKa = 3.34ILDD568 pKa = 3.94VGQSQALLLRR578 pKa = 11.84LPGHH582 pKa = 6.05VRR584 pKa = 11.84LLLDD588 pKa = 4.0GSGSASPRR596 pKa = 11.84FDD598 pKa = 3.44PGKK601 pKa = 10.62SLVAPVLVHH610 pKa = 6.7NDD612 pKa = 3.29APRR615 pKa = 11.84LTAVLNSHH623 pKa = 7.66PDD625 pKa = 3.56LDD627 pKa = 4.14HH628 pKa = 6.79MGGLLYY634 pKa = 10.25ILRR637 pKa = 11.84HH638 pKa = 5.71FRR640 pKa = 11.84TQGLFDD646 pKa = 3.64NGKK649 pKa = 8.32NAKK652 pKa = 9.63GKK654 pKa = 9.17WGEE657 pKa = 3.89LWDD660 pKa = 3.89LTRR663 pKa = 11.84QKK665 pKa = 10.61HH666 pKa = 4.57AARR669 pKa = 11.84PLRR672 pKa = 11.84QGDD675 pKa = 3.9TLVLGEE681 pKa = 4.7PEE683 pKa = 3.7NGLRR687 pKa = 11.84LEE689 pKa = 4.17ILHH692 pKa = 6.61PPRR695 pKa = 11.84HH696 pKa = 6.29SEE698 pKa = 4.56DD699 pKa = 2.58KK700 pKa = 10.36WRR702 pKa = 11.84GNNASLVARR711 pKa = 11.84LLHH714 pKa = 6.22HH715 pKa = 6.45EE716 pKa = 4.39RR717 pKa = 11.84GLALFMGDD725 pKa = 3.21AEE727 pKa = 4.39RR728 pKa = 11.84PVLQRR733 pKa = 11.84LLAEE737 pKa = 4.52GLDD740 pKa = 3.62VRR742 pKa = 11.84AEE744 pKa = 4.0VLIAPHH750 pKa = 6.55HH751 pKa = 5.82GSYY754 pKa = 10.03TGFLPQFFQAVRR766 pKa = 11.84PQLVVASCGSRR777 pKa = 11.84YY778 pKa = 10.02AYY780 pKa = 9.33PDD782 pKa = 2.93KK783 pKa = 10.88KK784 pKa = 10.53LRR786 pKa = 11.84EE787 pKa = 4.17WLAEE791 pKa = 3.88NHH793 pKa = 6.22ISLAYY798 pKa = 8.4TSRR801 pKa = 11.84DD802 pKa = 3.14GAVRR806 pKa = 11.84IAWQRR811 pKa = 11.84AGQGFSLPRR820 pKa = 11.84ISTARR825 pKa = 11.84KK826 pKa = 8.65QKK828 pKa = 10.78
MM1 pKa = 7.76RR2 pKa = 11.84HH3 pKa = 6.22PPLPAPLLWQSLLGFWILGILAVAWPVPSASCAFLLFLADD43 pKa = 3.36ARR45 pKa = 11.84LWRR48 pKa = 11.84AKK50 pKa = 10.77GIFAALAVLLAGVLAGHH67 pKa = 7.22LTTGKK72 pKa = 8.18MWRR75 pKa = 11.84DD76 pKa = 3.44HH77 pKa = 6.32TPTPVAWLDD86 pKa = 3.62TAPKK90 pKa = 10.3SMRR93 pKa = 11.84ICGQIQEE100 pKa = 4.39VRR102 pKa = 11.84GLPDD106 pKa = 2.93NRR108 pKa = 11.84IRR110 pKa = 11.84MLLADD115 pKa = 4.02VRR117 pKa = 11.84PHH119 pKa = 6.47GGAEE123 pKa = 3.96KK124 pKa = 10.52SLSGLMEE131 pKa = 4.11WTWQNPSAYY140 pKa = 8.68PLPGQSACLSRR151 pKa = 11.84RR152 pKa = 11.84PQAMQGFANPGLPDD166 pKa = 4.01RR167 pKa = 11.84DD168 pKa = 3.52VVIQSVRR175 pKa = 11.84NVRR178 pKa = 11.84WRR180 pKa = 11.84IWSQGEE186 pKa = 4.12SGQPRR191 pKa = 11.84IDD193 pKa = 3.75GQPSLSARR201 pKa = 11.84WRR203 pKa = 11.84EE204 pKa = 3.97SLRR207 pKa = 11.84QGFFTVLSPDD217 pKa = 3.5AEE219 pKa = 4.16HH220 pKa = 7.26FPRR223 pKa = 11.84QKK225 pKa = 10.64LPQGKK230 pKa = 9.82AILVALLFGDD240 pKa = 4.4RR241 pKa = 11.84QFLDD245 pKa = 3.36NQTIAGFSMAALGHH259 pKa = 5.87SLALSGQHH267 pKa = 6.35LAVAGLAGILLVVVVARR284 pKa = 11.84AYY286 pKa = 9.86PGIYY290 pKa = 9.79LCRR293 pKa = 11.84TRR295 pKa = 11.84TVWVMSASVPPALAYY310 pKa = 10.37LWLGNAPPSLVRR322 pKa = 11.84ATIMLCILALWLLRR336 pKa = 11.84RR337 pKa = 11.84RR338 pKa = 11.84VHH340 pKa = 5.12TTLDD344 pKa = 3.73VLCSALFCITLFSPAAVFDD363 pKa = 3.81TGLQLSAICVAVIGMSMPWLRR384 pKa = 11.84RR385 pKa = 11.84LLPRR389 pKa = 11.84PHH391 pKa = 6.91PAEE394 pKa = 3.99HH395 pKa = 6.4AARR398 pKa = 11.84WRR400 pKa = 11.84SLLQDD405 pKa = 4.43ALWSLQRR412 pKa = 11.84IFALSLIIQTVLLPVGILLFGNTGHH437 pKa = 7.51WFPLNVLWLPLVNLLVLPGAALGLLAGACGWDD469 pKa = 3.18AAARR473 pKa = 11.84AILEE477 pKa = 4.34YY478 pKa = 10.93ASLPCQWLADD488 pKa = 3.83ALAWLARR495 pKa = 11.84QNALEE500 pKa = 4.23SPVVLRR506 pKa = 11.84PHH508 pKa = 6.04WTALPAFAALTAAAALRR525 pKa = 11.84GGRR528 pKa = 11.84LRR530 pKa = 11.84LPPAGRR536 pKa = 11.84RR537 pKa = 11.84LLLAGALLLCAGPLLRR553 pKa = 11.84AGEE556 pKa = 4.13RR557 pKa = 11.84LSPEE561 pKa = 3.61VRR563 pKa = 11.84LDD565 pKa = 3.34ILDD568 pKa = 3.94VGQSQALLLRR578 pKa = 11.84LPGHH582 pKa = 6.05VRR584 pKa = 11.84LLLDD588 pKa = 4.0GSGSASPRR596 pKa = 11.84FDD598 pKa = 3.44PGKK601 pKa = 10.62SLVAPVLVHH610 pKa = 6.7NDD612 pKa = 3.29APRR615 pKa = 11.84LTAVLNSHH623 pKa = 7.66PDD625 pKa = 3.56LDD627 pKa = 4.14HH628 pKa = 6.79MGGLLYY634 pKa = 10.25ILRR637 pKa = 11.84HH638 pKa = 5.71FRR640 pKa = 11.84TQGLFDD646 pKa = 3.64NGKK649 pKa = 8.32NAKK652 pKa = 9.63GKK654 pKa = 9.17WGEE657 pKa = 3.89LWDD660 pKa = 3.89LTRR663 pKa = 11.84QKK665 pKa = 10.61HH666 pKa = 4.57AARR669 pKa = 11.84PLRR672 pKa = 11.84QGDD675 pKa = 3.9TLVLGEE681 pKa = 4.7PEE683 pKa = 3.7NGLRR687 pKa = 11.84LEE689 pKa = 4.17ILHH692 pKa = 6.61PPRR695 pKa = 11.84HH696 pKa = 6.29SEE698 pKa = 4.56DD699 pKa = 2.58KK700 pKa = 10.36WRR702 pKa = 11.84GNNASLVARR711 pKa = 11.84LLHH714 pKa = 6.22HH715 pKa = 6.45EE716 pKa = 4.39RR717 pKa = 11.84GLALFMGDD725 pKa = 3.21AEE727 pKa = 4.39RR728 pKa = 11.84PVLQRR733 pKa = 11.84LLAEE737 pKa = 4.52GLDD740 pKa = 3.62VRR742 pKa = 11.84AEE744 pKa = 4.0VLIAPHH750 pKa = 6.55HH751 pKa = 5.82GSYY754 pKa = 10.03TGFLPQFFQAVRR766 pKa = 11.84PQLVVASCGSRR777 pKa = 11.84YY778 pKa = 10.02AYY780 pKa = 9.33PDD782 pKa = 2.93KK783 pKa = 10.88KK784 pKa = 10.53LRR786 pKa = 11.84EE787 pKa = 4.17WLAEE791 pKa = 3.88NHH793 pKa = 6.22ISLAYY798 pKa = 8.4TSRR801 pKa = 11.84DD802 pKa = 3.14GAVRR806 pKa = 11.84IAWQRR811 pKa = 11.84AGQGFSLPRR820 pKa = 11.84ISTARR825 pKa = 11.84KK826 pKa = 8.65QKK828 pKa = 10.78
Molecular weight: 90.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
758626 |
26 |
3022 |
319.7 |
35.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.204 ± 0.069 | 1.657 ± 0.026 |
5.071 ± 0.037 | 5.967 ± 0.049 |
3.827 ± 0.031 | 8.152 ± 0.048 |
2.337 ± 0.027 | 4.431 ± 0.046 |
3.862 ± 0.042 | 11.147 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.658 ± 0.027 | 2.853 ± 0.036 |
5.245 ± 0.045 | 3.463 ± 0.027 |
7.046 ± 0.054 | 5.592 ± 0.031 |
4.752 ± 0.049 | 7.033 ± 0.043 |
1.317 ± 0.02 | 2.389 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
