
Grasshopper associated circular virus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykolovirus; Gemykolovirus easlu1
Average proteome isoelectric point is 7.92
Get precalculated fractions of proteins
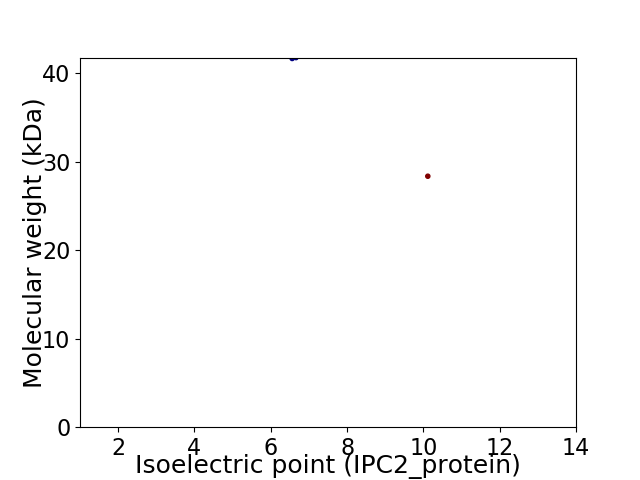
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BP63|A0A346BP63_9VIRU Putative capsid protein OS=Grasshopper associated circular virus 1 OX=2293293 PE=4 SV=1
MM1 pKa = 6.92VRR3 pKa = 11.84KK4 pKa = 9.98FKK6 pKa = 10.86LDD8 pKa = 3.21NVNYY12 pKa = 10.36VLLTYY17 pKa = 10.46SDD19 pKa = 4.65CPNDD23 pKa = 4.17FDD25 pKa = 4.18PQLIIDD31 pKa = 4.19AVVGTGAVYY40 pKa = 10.28RR41 pKa = 11.84LGRR44 pKa = 11.84EE45 pKa = 3.54LHH47 pKa = 6.92ANGKK51 pKa = 7.8PHH53 pKa = 6.25FHH55 pKa = 7.14CFVQWAEE62 pKa = 4.28PFSHH66 pKa = 7.62PDD68 pKa = 2.97AGSLFFVGGRR78 pKa = 11.84RR79 pKa = 11.84ANIKK83 pKa = 10.19RR84 pKa = 11.84FSANPGRR91 pKa = 11.84RR92 pKa = 11.84WDD94 pKa = 3.79YY95 pKa = 8.01VGKK98 pKa = 10.45YY99 pKa = 9.04AGHH102 pKa = 6.91KK103 pKa = 9.66AGHH106 pKa = 6.62FIIGDD111 pKa = 3.44QCDD114 pKa = 3.52RR115 pKa = 11.84PGGDD119 pKa = 3.56KK120 pKa = 10.72DD121 pKa = 4.04DD122 pKa = 4.47SEE124 pKa = 4.46RR125 pKa = 11.84TQTDD129 pKa = 2.27IWSEE133 pKa = 3.94IINAEE138 pKa = 4.1TEE140 pKa = 4.25SEE142 pKa = 4.24FFEE145 pKa = 4.71KK146 pKa = 10.56LASLAPKK153 pKa = 10.27QLGCNFGSLKK163 pKa = 10.7LYY165 pKa = 10.78ADD167 pKa = 3.64WRR169 pKa = 11.84YY170 pKa = 10.03KK171 pKa = 10.78PPVTEE176 pKa = 4.47YY177 pKa = 10.35ISPSGNFYY185 pKa = 8.47PTTALADD192 pKa = 3.62WEE194 pKa = 4.3RR195 pKa = 11.84EE196 pKa = 3.9QLNSEE201 pKa = 3.97MSACLRR207 pKa = 11.84GGPLATPSPSASRR220 pKa = 11.84AGPHH224 pKa = 6.43RR225 pKa = 11.84LLPKK229 pKa = 10.5HH230 pKa = 5.54MLILEE235 pKa = 4.88FIRR238 pKa = 11.84PRR240 pKa = 11.84GLVLFGATRR249 pKa = 11.84LGKK252 pKa = 7.65TVWARR257 pKa = 11.84SLGTHH262 pKa = 6.35SYY264 pKa = 10.87FGGLFNMEE272 pKa = 4.71LFAEE276 pKa = 4.55EE277 pKa = 3.3ARR279 pKa = 11.84YY280 pKa = 9.95AIFDD284 pKa = 5.01DD285 pKa = 3.55ISGGFGFFPSYY296 pKa = 10.73KK297 pKa = 9.81LWLGGQFEE305 pKa = 4.69FSVTDD310 pKa = 3.54KK311 pKa = 11.09YY312 pKa = 10.68KK313 pKa = 10.67HH314 pKa = 5.7KK315 pKa = 11.09RR316 pKa = 11.84NVKK319 pKa = 8.0WGKK322 pKa = 8.34PAIWLCNTDD331 pKa = 5.46PRR333 pKa = 11.84LDD335 pKa = 3.32WYY337 pKa = 11.08KK338 pKa = 10.71PGTGPDD344 pKa = 3.99FEE346 pKa = 4.64WMEE349 pKa = 4.2NNCDD353 pKa = 3.33FVEE356 pKa = 3.99ITEE359 pKa = 4.77PIFRR363 pKa = 11.84ANTTT367 pKa = 3.29
MM1 pKa = 6.92VRR3 pKa = 11.84KK4 pKa = 9.98FKK6 pKa = 10.86LDD8 pKa = 3.21NVNYY12 pKa = 10.36VLLTYY17 pKa = 10.46SDD19 pKa = 4.65CPNDD23 pKa = 4.17FDD25 pKa = 4.18PQLIIDD31 pKa = 4.19AVVGTGAVYY40 pKa = 10.28RR41 pKa = 11.84LGRR44 pKa = 11.84EE45 pKa = 3.54LHH47 pKa = 6.92ANGKK51 pKa = 7.8PHH53 pKa = 6.25FHH55 pKa = 7.14CFVQWAEE62 pKa = 4.28PFSHH66 pKa = 7.62PDD68 pKa = 2.97AGSLFFVGGRR78 pKa = 11.84RR79 pKa = 11.84ANIKK83 pKa = 10.19RR84 pKa = 11.84FSANPGRR91 pKa = 11.84RR92 pKa = 11.84WDD94 pKa = 3.79YY95 pKa = 8.01VGKK98 pKa = 10.45YY99 pKa = 9.04AGHH102 pKa = 6.91KK103 pKa = 9.66AGHH106 pKa = 6.62FIIGDD111 pKa = 3.44QCDD114 pKa = 3.52RR115 pKa = 11.84PGGDD119 pKa = 3.56KK120 pKa = 10.72DD121 pKa = 4.04DD122 pKa = 4.47SEE124 pKa = 4.46RR125 pKa = 11.84TQTDD129 pKa = 2.27IWSEE133 pKa = 3.94IINAEE138 pKa = 4.1TEE140 pKa = 4.25SEE142 pKa = 4.24FFEE145 pKa = 4.71KK146 pKa = 10.56LASLAPKK153 pKa = 10.27QLGCNFGSLKK163 pKa = 10.7LYY165 pKa = 10.78ADD167 pKa = 3.64WRR169 pKa = 11.84YY170 pKa = 10.03KK171 pKa = 10.78PPVTEE176 pKa = 4.47YY177 pKa = 10.35ISPSGNFYY185 pKa = 8.47PTTALADD192 pKa = 3.62WEE194 pKa = 4.3RR195 pKa = 11.84EE196 pKa = 3.9QLNSEE201 pKa = 3.97MSACLRR207 pKa = 11.84GGPLATPSPSASRR220 pKa = 11.84AGPHH224 pKa = 6.43RR225 pKa = 11.84LLPKK229 pKa = 10.5HH230 pKa = 5.54MLILEE235 pKa = 4.88FIRR238 pKa = 11.84PRR240 pKa = 11.84GLVLFGATRR249 pKa = 11.84LGKK252 pKa = 7.65TVWARR257 pKa = 11.84SLGTHH262 pKa = 6.35SYY264 pKa = 10.87FGGLFNMEE272 pKa = 4.71LFAEE276 pKa = 4.55EE277 pKa = 3.3ARR279 pKa = 11.84YY280 pKa = 9.95AIFDD284 pKa = 5.01DD285 pKa = 3.55ISGGFGFFPSYY296 pKa = 10.73KK297 pKa = 9.81LWLGGQFEE305 pKa = 4.69FSVTDD310 pKa = 3.54KK311 pKa = 11.09YY312 pKa = 10.68KK313 pKa = 10.67HH314 pKa = 5.7KK315 pKa = 11.09RR316 pKa = 11.84NVKK319 pKa = 8.0WGKK322 pKa = 8.34PAIWLCNTDD331 pKa = 5.46PRR333 pKa = 11.84LDD335 pKa = 3.32WYY337 pKa = 11.08KK338 pKa = 10.71PGTGPDD344 pKa = 3.99FEE346 pKa = 4.64WMEE349 pKa = 4.2NNCDD353 pKa = 3.33FVEE356 pKa = 3.99ITEE359 pKa = 4.77PIFRR363 pKa = 11.84ANTTT367 pKa = 3.29
Molecular weight: 41.66 kDa
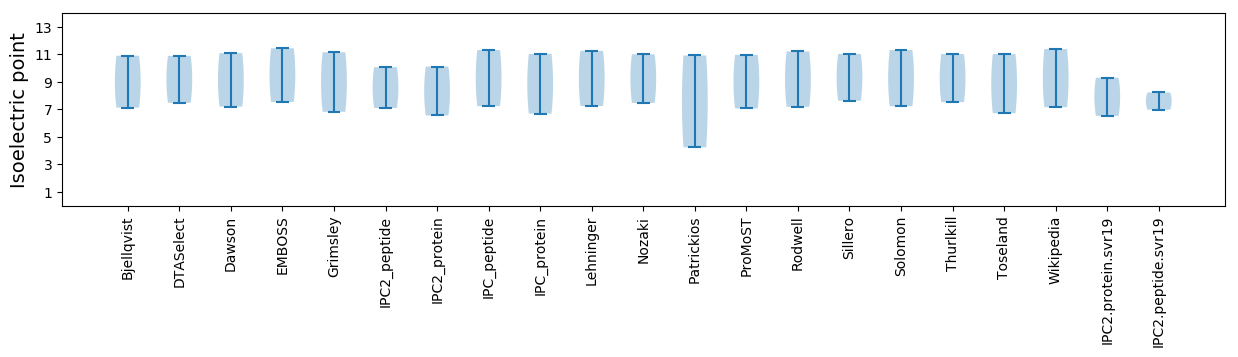
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BP63|A0A346BP63_9VIRU Putative capsid protein OS=Grasshopper associated circular virus 1 OX=2293293 PE=4 SV=1
MM1 pKa = 7.99AYY3 pKa = 9.8RR4 pKa = 11.84RR5 pKa = 11.84STRR8 pKa = 11.84RR9 pKa = 11.84YY10 pKa = 7.51AARR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84PTRR18 pKa = 11.84KK19 pKa = 8.63RR20 pKa = 11.84SYY22 pKa = 9.16PSRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84YY28 pKa = 7.07TGKK31 pKa = 9.99RR32 pKa = 11.84RR33 pKa = 11.84STSRR37 pKa = 11.84KK38 pKa = 5.12MTTKK42 pKa = 10.25RR43 pKa = 11.84VLNVTSRR50 pKa = 11.84KK51 pKa = 9.89KK52 pKa = 10.33QDD54 pKa = 2.85TMQNLTNYY62 pKa = 9.68SAASVSGGTQFYY74 pKa = 10.58SDD76 pKa = 3.43NTIISGGTLFASLWCPTARR95 pKa = 11.84TNHH98 pKa = 6.05AQLQAKK104 pKa = 7.75DD105 pKa = 3.48QPEE108 pKa = 4.02ILGGLFDD115 pKa = 4.04KK116 pKa = 10.33ATRR119 pKa = 11.84TASTCYY125 pKa = 9.67MRR127 pKa = 11.84GLKK130 pKa = 10.0EE131 pKa = 4.21KK132 pKa = 10.51IQIQTNTGVAWQWRR146 pKa = 11.84RR147 pKa = 11.84ICFTAKK153 pKa = 10.11GGSYY157 pKa = 10.22YY158 pKa = 11.27NGMTDD163 pKa = 3.1NAKK166 pKa = 10.23LVAFAGTGWARR177 pKa = 11.84VTSNWLFSDD186 pKa = 4.22PSKK189 pKa = 10.84QALFGPLFRR198 pKa = 11.84GQEE201 pKa = 4.17GVDD204 pKa = 2.85WRR206 pKa = 11.84NYY208 pKa = 9.36FSAPLDD214 pKa = 3.52THH216 pKa = 6.9RR217 pKa = 11.84MNVKK221 pKa = 9.69YY222 pKa = 10.18DD223 pKa = 3.32KK224 pKa = 10.87TVIIQSGNDD233 pKa = 3.21SGVMRR238 pKa = 11.84NYY240 pKa = 10.32SRR242 pKa = 11.84SPWTPTAA249 pKa = 3.83
MM1 pKa = 7.99AYY3 pKa = 9.8RR4 pKa = 11.84RR5 pKa = 11.84STRR8 pKa = 11.84RR9 pKa = 11.84YY10 pKa = 7.51AARR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84PTRR18 pKa = 11.84KK19 pKa = 8.63RR20 pKa = 11.84SYY22 pKa = 9.16PSRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84YY28 pKa = 7.07TGKK31 pKa = 9.99RR32 pKa = 11.84RR33 pKa = 11.84STSRR37 pKa = 11.84KK38 pKa = 5.12MTTKK42 pKa = 10.25RR43 pKa = 11.84VLNVTSRR50 pKa = 11.84KK51 pKa = 9.89KK52 pKa = 10.33QDD54 pKa = 2.85TMQNLTNYY62 pKa = 9.68SAASVSGGTQFYY74 pKa = 10.58SDD76 pKa = 3.43NTIISGGTLFASLWCPTARR95 pKa = 11.84TNHH98 pKa = 6.05AQLQAKK104 pKa = 7.75DD105 pKa = 3.48QPEE108 pKa = 4.02ILGGLFDD115 pKa = 4.04KK116 pKa = 10.33ATRR119 pKa = 11.84TASTCYY125 pKa = 9.67MRR127 pKa = 11.84GLKK130 pKa = 10.0EE131 pKa = 4.21KK132 pKa = 10.51IQIQTNTGVAWQWRR146 pKa = 11.84RR147 pKa = 11.84ICFTAKK153 pKa = 10.11GGSYY157 pKa = 10.22YY158 pKa = 11.27NGMTDD163 pKa = 3.1NAKK166 pKa = 10.23LVAFAGTGWARR177 pKa = 11.84VTSNWLFSDD186 pKa = 4.22PSKK189 pKa = 10.84QALFGPLFRR198 pKa = 11.84GQEE201 pKa = 4.17GVDD204 pKa = 2.85WRR206 pKa = 11.84NYY208 pKa = 9.36FSAPLDD214 pKa = 3.52THH216 pKa = 6.9RR217 pKa = 11.84MNVKK221 pKa = 9.69YY222 pKa = 10.18DD223 pKa = 3.32KK224 pKa = 10.87TVIIQSGNDD233 pKa = 3.21SGVMRR238 pKa = 11.84NYY240 pKa = 10.32SRR242 pKa = 11.84SPWTPTAA249 pKa = 3.83
Molecular weight: 28.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
616 |
249 |
367 |
308.0 |
35.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.468 ± 0.377 | 1.623 ± 0.279 |
5.195 ± 0.787 | 3.896 ± 1.797 |
5.844 ± 1.489 | 8.604 ± 0.65 |
1.948 ± 0.764 | 3.896 ± 0.456 |
5.682 ± 0.229 | 6.981 ± 1.175 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.948 ± 0.576 | 4.708 ± 0.343 |
5.357 ± 1.163 | 3.084 ± 1.158 |
8.279 ± 1.98 | 6.981 ± 1.238 |
7.305 ± 2.362 | 4.058 ± 0.028 |
2.922 ± 0.074 | 4.221 ± 0.4 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
