
Drosophila willistoni (Fruit fly)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Diptera; Brachycera; Muscomorpha; Eremoneura; Cyclorrhapha; Schizophora; Acalyptratae;
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
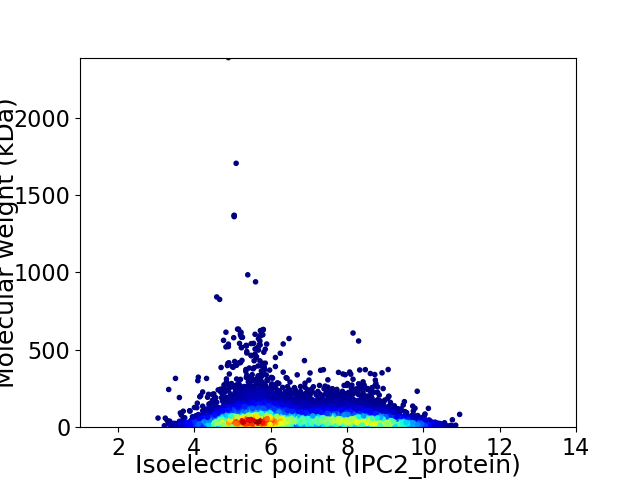
Virtual 2D-PAGE plot for 14820 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B4MTW5|B4MTW5_DROWI BEN domain-containing protein OS=Drosophila willistoni OX=7260 GN=Dwil\GK23918 PE=4 SV=2
MM1 pKa = 7.31FNKK4 pKa = 10.22SYY6 pKa = 11.1TNIILASTLLLCLITLPVSQADD28 pKa = 3.74LMCYY32 pKa = 10.18VCDD35 pKa = 5.0DD36 pKa = 4.38CATMPKK42 pKa = 10.1DD43 pKa = 4.81APLLACNEE51 pKa = 4.28DD52 pKa = 3.71FFNPGGSTEE61 pKa = 4.14ASTVTTTTPASTTEE75 pKa = 4.29AVQSTTPIEE84 pKa = 4.11TSPMTIASTTEE95 pKa = 3.73EE96 pKa = 4.43SSTDD100 pKa = 3.52LASTTTEE107 pKa = 4.12EE108 pKa = 4.38VTSTSTSEE116 pKa = 3.81QSTEE120 pKa = 3.97STEE123 pKa = 4.6APIPTPNTAGPVEE136 pKa = 4.5TTTGVPTPPQEE147 pKa = 5.19DD148 pKa = 4.06LDD150 pKa = 4.23AVPASAANTTRR161 pKa = 11.84LLQIGEE167 pKa = 4.25EE168 pKa = 4.25SSSNVTPLAIRR179 pKa = 11.84QRR181 pKa = 11.84RR182 pKa = 11.84ALINTDD188 pKa = 3.47YY189 pKa = 10.98SYY191 pKa = 11.43HH192 pKa = 6.95CYY194 pKa = 9.92IVQKK198 pKa = 10.11SINGSTVTDD207 pKa = 4.24RR208 pKa = 11.84GCSRR212 pKa = 11.84VSTYY216 pKa = 10.95QSVCGLLMEE225 pKa = 5.06QNNNTQLSNCDD236 pKa = 3.59PCSMNACNGSSLLQNSLLVTFLMAAIAALMQANVLYY272 pKa = 10.84KK273 pKa = 10.46KK274 pKa = 9.65MRR276 pKa = 11.84LISLLLITLICAGATIWSANALQCYY301 pKa = 8.68EE302 pKa = 4.81CIGKK306 pKa = 9.12EE307 pKa = 3.9CDD309 pKa = 3.39EE310 pKa = 4.31LTEE313 pKa = 4.05EE314 pKa = 4.82HH315 pKa = 6.91LVSCNVDD322 pKa = 3.18NDD324 pKa = 4.13PVTTPSDD331 pKa = 3.72EE332 pKa = 4.26TDD334 pKa = 3.5STDD337 pKa = 3.33DD338 pKa = 3.68TTDD341 pKa = 2.74KK342 pKa = 10.08STPSATSSTTNGVTTSSPGTTTGTTGTTDD371 pKa = 3.6ADD373 pKa = 3.99TTTTTDD379 pKa = 2.8EE380 pKa = 5.29GYY382 pKa = 10.88SSTIEE387 pKa = 4.39DD388 pKa = 3.98EE389 pKa = 4.82SSVSTDD395 pKa = 3.92DD396 pKa = 5.49DD397 pKa = 4.41SSSSNTTDD405 pKa = 4.63DD406 pKa = 4.49DD407 pKa = 4.74SSTNTEE413 pKa = 4.13SSTSDD418 pKa = 3.03TTEE421 pKa = 4.65DD422 pKa = 4.06DD423 pKa = 3.93SSSSSNDD430 pKa = 3.21DD431 pKa = 3.46SSSSSTTEE439 pKa = 3.87STSEE443 pKa = 3.89SSPNTEE449 pKa = 4.07SSTSDD454 pKa = 3.03TTEE457 pKa = 4.65DD458 pKa = 4.06DD459 pKa = 3.93SSSSSNDD466 pKa = 3.25DD467 pKa = 3.3SSSSITTEE475 pKa = 3.9SSSEE479 pKa = 3.99SSPNAEE485 pKa = 4.24SSTSDD490 pKa = 3.07TTEE493 pKa = 4.67DD494 pKa = 4.14DD495 pKa = 3.91SSSSTNADD503 pKa = 3.08SSSSNTTDD511 pKa = 3.37EE512 pKa = 4.84DD513 pKa = 4.32SSTNTEE519 pKa = 4.09SSTSDD524 pKa = 3.03TTEE527 pKa = 4.65DD528 pKa = 4.06DD529 pKa = 3.93SSSSSNDD536 pKa = 3.04EE537 pKa = 4.06SSSSITTEE545 pKa = 4.03SASEE549 pKa = 4.01SSPNAEE555 pKa = 4.24SSTSDD560 pKa = 3.07TTEE563 pKa = 4.65DD564 pKa = 4.06DD565 pKa = 3.93SSSSSNDD572 pKa = 3.24DD573 pKa = 3.31SSSTIVTEE581 pKa = 4.33SEE583 pKa = 4.38SPSQSSIDD591 pKa = 3.96AEE593 pKa = 4.49SSTSGIIEE601 pKa = 4.95DD602 pKa = 4.74DD603 pKa = 3.7SSSASSGASEE613 pKa = 4.32SSSASSEE620 pKa = 4.25TSSSGAEE627 pKa = 3.91TTVGSTEE634 pKa = 3.96APVTSRR640 pKa = 11.84AKK642 pKa = 10.25RR643 pKa = 11.84SLAARR648 pKa = 11.84ATDD651 pKa = 3.91RR652 pKa = 11.84LLQNYY657 pKa = 9.21RR658 pKa = 11.84SYY660 pKa = 11.77ARR662 pKa = 11.84YY663 pKa = 9.59RR664 pKa = 11.84RR665 pKa = 11.84DD666 pKa = 2.93AEE668 pKa = 4.15GQIRR672 pKa = 11.84TTACYY677 pKa = 10.44SIVKK681 pKa = 10.35DD682 pKa = 3.58NVIQRR687 pKa = 11.84GCIRR691 pKa = 11.84IPYY694 pKa = 9.21GKK696 pKa = 9.79GGCEE700 pKa = 3.41AVRR703 pKa = 11.84IEE705 pKa = 4.36VNLAADD711 pKa = 4.28SIGDD715 pKa = 3.78EE716 pKa = 4.34CDD718 pKa = 2.95VCQMDD723 pKa = 4.02KK724 pKa = 11.15CNSSTSLRR732 pKa = 11.84LSLGLMLLGLITFGLRR748 pKa = 11.84SLL750 pKa = 4.12
MM1 pKa = 7.31FNKK4 pKa = 10.22SYY6 pKa = 11.1TNIILASTLLLCLITLPVSQADD28 pKa = 3.74LMCYY32 pKa = 10.18VCDD35 pKa = 5.0DD36 pKa = 4.38CATMPKK42 pKa = 10.1DD43 pKa = 4.81APLLACNEE51 pKa = 4.28DD52 pKa = 3.71FFNPGGSTEE61 pKa = 4.14ASTVTTTTPASTTEE75 pKa = 4.29AVQSTTPIEE84 pKa = 4.11TSPMTIASTTEE95 pKa = 3.73EE96 pKa = 4.43SSTDD100 pKa = 3.52LASTTTEE107 pKa = 4.12EE108 pKa = 4.38VTSTSTSEE116 pKa = 3.81QSTEE120 pKa = 3.97STEE123 pKa = 4.6APIPTPNTAGPVEE136 pKa = 4.5TTTGVPTPPQEE147 pKa = 5.19DD148 pKa = 4.06LDD150 pKa = 4.23AVPASAANTTRR161 pKa = 11.84LLQIGEE167 pKa = 4.25EE168 pKa = 4.25SSSNVTPLAIRR179 pKa = 11.84QRR181 pKa = 11.84RR182 pKa = 11.84ALINTDD188 pKa = 3.47YY189 pKa = 10.98SYY191 pKa = 11.43HH192 pKa = 6.95CYY194 pKa = 9.92IVQKK198 pKa = 10.11SINGSTVTDD207 pKa = 4.24RR208 pKa = 11.84GCSRR212 pKa = 11.84VSTYY216 pKa = 10.95QSVCGLLMEE225 pKa = 5.06QNNNTQLSNCDD236 pKa = 3.59PCSMNACNGSSLLQNSLLVTFLMAAIAALMQANVLYY272 pKa = 10.84KK273 pKa = 10.46KK274 pKa = 9.65MRR276 pKa = 11.84LISLLLITLICAGATIWSANALQCYY301 pKa = 8.68EE302 pKa = 4.81CIGKK306 pKa = 9.12EE307 pKa = 3.9CDD309 pKa = 3.39EE310 pKa = 4.31LTEE313 pKa = 4.05EE314 pKa = 4.82HH315 pKa = 6.91LVSCNVDD322 pKa = 3.18NDD324 pKa = 4.13PVTTPSDD331 pKa = 3.72EE332 pKa = 4.26TDD334 pKa = 3.5STDD337 pKa = 3.33DD338 pKa = 3.68TTDD341 pKa = 2.74KK342 pKa = 10.08STPSATSSTTNGVTTSSPGTTTGTTGTTDD371 pKa = 3.6ADD373 pKa = 3.99TTTTTDD379 pKa = 2.8EE380 pKa = 5.29GYY382 pKa = 10.88SSTIEE387 pKa = 4.39DD388 pKa = 3.98EE389 pKa = 4.82SSVSTDD395 pKa = 3.92DD396 pKa = 5.49DD397 pKa = 4.41SSSSNTTDD405 pKa = 4.63DD406 pKa = 4.49DD407 pKa = 4.74SSTNTEE413 pKa = 4.13SSTSDD418 pKa = 3.03TTEE421 pKa = 4.65DD422 pKa = 4.06DD423 pKa = 3.93SSSSSNDD430 pKa = 3.21DD431 pKa = 3.46SSSSSTTEE439 pKa = 3.87STSEE443 pKa = 3.89SSPNTEE449 pKa = 4.07SSTSDD454 pKa = 3.03TTEE457 pKa = 4.65DD458 pKa = 4.06DD459 pKa = 3.93SSSSSNDD466 pKa = 3.25DD467 pKa = 3.3SSSSITTEE475 pKa = 3.9SSSEE479 pKa = 3.99SSPNAEE485 pKa = 4.24SSTSDD490 pKa = 3.07TTEE493 pKa = 4.67DD494 pKa = 4.14DD495 pKa = 3.91SSSSTNADD503 pKa = 3.08SSSSNTTDD511 pKa = 3.37EE512 pKa = 4.84DD513 pKa = 4.32SSTNTEE519 pKa = 4.09SSTSDD524 pKa = 3.03TTEE527 pKa = 4.65DD528 pKa = 4.06DD529 pKa = 3.93SSSSSNDD536 pKa = 3.04EE537 pKa = 4.06SSSSITTEE545 pKa = 4.03SASEE549 pKa = 4.01SSPNAEE555 pKa = 4.24SSTSDD560 pKa = 3.07TTEE563 pKa = 4.65DD564 pKa = 4.06DD565 pKa = 3.93SSSSSNDD572 pKa = 3.24DD573 pKa = 3.31SSSTIVTEE581 pKa = 4.33SEE583 pKa = 4.38SPSQSSIDD591 pKa = 3.96AEE593 pKa = 4.49SSTSGIIEE601 pKa = 4.95DD602 pKa = 4.74DD603 pKa = 3.7SSSASSGASEE613 pKa = 4.32SSSASSEE620 pKa = 4.25TSSSGAEE627 pKa = 3.91TTVGSTEE634 pKa = 3.96APVTSRR640 pKa = 11.84AKK642 pKa = 10.25RR643 pKa = 11.84SLAARR648 pKa = 11.84ATDD651 pKa = 3.91RR652 pKa = 11.84LLQNYY657 pKa = 9.21RR658 pKa = 11.84SYY660 pKa = 11.77ARR662 pKa = 11.84YY663 pKa = 9.59RR664 pKa = 11.84RR665 pKa = 11.84DD666 pKa = 2.93AEE668 pKa = 4.15GQIRR672 pKa = 11.84TTACYY677 pKa = 10.44SIVKK681 pKa = 10.35DD682 pKa = 3.58NVIQRR687 pKa = 11.84GCIRR691 pKa = 11.84IPYY694 pKa = 9.21GKK696 pKa = 9.79GGCEE700 pKa = 3.41AVRR703 pKa = 11.84IEE705 pKa = 4.36VNLAADD711 pKa = 4.28SIGDD715 pKa = 3.78EE716 pKa = 4.34CDD718 pKa = 2.95VCQMDD723 pKa = 4.02KK724 pKa = 11.15CNSSTSLRR732 pKa = 11.84LSLGLMLLGLITFGLRR748 pKa = 11.84SLL750 pKa = 4.12
Molecular weight: 78.08 kDa
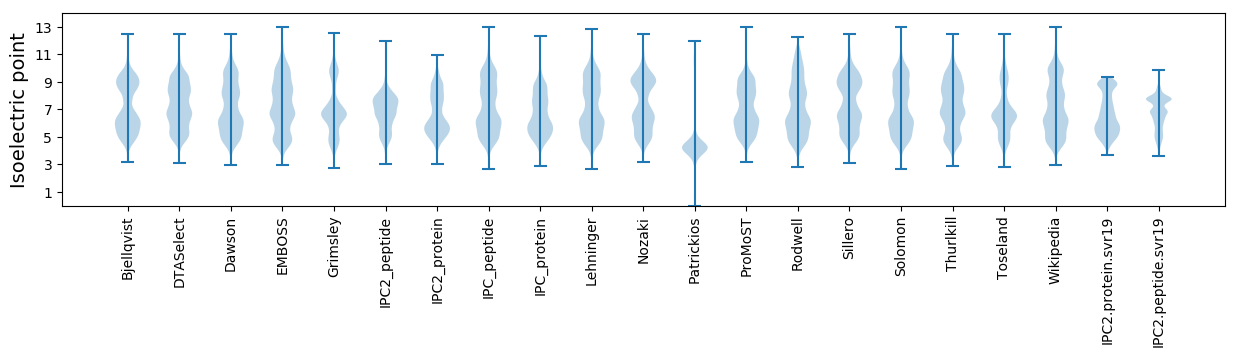
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B4NJF2|B4NJF2_DROWI Serine/threonine-protein phosphatase OS=Drosophila willistoni OX=7260 GN=Dwil\GK12844 PE=3 SV=1
MM1 pKa = 7.71PKK3 pKa = 10.26SLLDD7 pKa = 3.86YY8 pKa = 10.8SPHH11 pKa = 6.65GYY13 pKa = 10.23DD14 pKa = 2.71WLFLFVVFFIFAGGLGNILVCLAVALDD41 pKa = 3.71RR42 pKa = 11.84KK43 pKa = 9.47LQNVTNYY50 pKa = 10.16FLFSLAIADD59 pKa = 4.27LLVSLFVMPMGAIPAFLGYY78 pKa = 9.61WPLGFTWCNIYY89 pKa = 8.07VTCDD93 pKa = 3.17VLACSSSILHH103 pKa = 5.91MCFISLGRR111 pKa = 11.84YY112 pKa = 9.06LGIRR116 pKa = 11.84NPLGSRR122 pKa = 11.84HH123 pKa = 6.34RR124 pKa = 11.84STKK127 pKa = 9.86RR128 pKa = 11.84LAGIKK133 pKa = 9.64IAIVWVMAMMVSSSITVLGLVNKK156 pKa = 9.48HH157 pKa = 6.27NIMPEE162 pKa = 3.8PNICMINNRR171 pKa = 11.84AFWVFGSLVAFYY183 pKa = 10.53IPMLMMVTTYY193 pKa = 11.64ALTIPLLRR201 pKa = 11.84KK202 pKa = 9.14KK203 pKa = 10.66ARR205 pKa = 11.84FAAEE209 pKa = 4.06HH210 pKa = 6.84PEE212 pKa = 4.03SEE214 pKa = 4.24LFRR217 pKa = 11.84RR218 pKa = 11.84LGGRR222 pKa = 11.84FTIRR226 pKa = 11.84PQHH229 pKa = 5.79SQQQLQMHH237 pKa = 6.74SSFSSNSNKK246 pKa = 9.24FLAMSDD252 pKa = 3.36SNRR255 pKa = 11.84NFNSSDD261 pKa = 3.18RR262 pKa = 11.84GGGRR266 pKa = 11.84GGDD269 pKa = 3.44NSTEE273 pKa = 3.96RR274 pKa = 11.84PLMQQRR280 pKa = 11.84TTSNRR285 pKa = 11.84SMGAGSVSFRR295 pKa = 11.84NVTNGGRR302 pKa = 11.84TAFGVGMGAGGRR314 pKa = 11.84SSFRR318 pKa = 11.84FAGGSILRR326 pKa = 11.84QSSTPTTTSTSLSSSCPSSTATSVRR351 pKa = 11.84GGRR354 pKa = 11.84SQQTSSFWRR363 pKa = 11.84KK364 pKa = 9.57HH365 pKa = 4.85GGNPNLMDD373 pKa = 3.28SHH375 pKa = 6.74PNRR378 pKa = 11.84RR379 pKa = 11.84ASVRR383 pKa = 11.84VSMSQPQLGYY393 pKa = 7.52PTNGNGGNGGGIGGNEE409 pKa = 4.26SPTAVTSSSCTTPGSISNLVNVATIQGQSMDD440 pKa = 3.38NHH442 pKa = 5.53QLEE445 pKa = 5.09HH446 pKa = 6.09GTTMTATITSAATTAASASARR467 pKa = 11.84ATTTTTGEE475 pKa = 4.1RR476 pKa = 11.84RR477 pKa = 11.84HH478 pKa = 6.08KK479 pKa = 10.53LRR481 pKa = 11.84PFKK484 pKa = 10.6FALNRR489 pKa = 11.84VATPTLNLRR498 pKa = 11.84FLNNRR503 pKa = 11.84NKK505 pKa = 10.44RR506 pKa = 11.84NSLSANAVATEE517 pKa = 3.99QKK519 pKa = 8.69ATKK522 pKa = 10.48VLGLVFFTFVLCWSPFFILNIIFAACPQCQVSEE555 pKa = 4.41HH556 pKa = 5.38VVNTCLWLGYY566 pKa = 10.09VSSTINPIIYY576 pKa = 9.08TIFNRR581 pKa = 11.84TFRR584 pKa = 11.84AAFIRR589 pKa = 11.84LLKK592 pKa = 10.36CNCEE596 pKa = 3.68RR597 pKa = 11.84SGRR600 pKa = 11.84PLRR603 pKa = 11.84FRR605 pKa = 11.84SVTEE609 pKa = 3.85GRR611 pKa = 11.84SALSLCAPSALPLAISFQGAPLMTPSTTNATPLSEE646 pKa = 4.21FRR648 pKa = 11.84GSYY651 pKa = 10.02TITDD655 pKa = 3.95DD656 pKa = 3.51EE657 pKa = 4.72CC658 pKa = 6.48
MM1 pKa = 7.71PKK3 pKa = 10.26SLLDD7 pKa = 3.86YY8 pKa = 10.8SPHH11 pKa = 6.65GYY13 pKa = 10.23DD14 pKa = 2.71WLFLFVVFFIFAGGLGNILVCLAVALDD41 pKa = 3.71RR42 pKa = 11.84KK43 pKa = 9.47LQNVTNYY50 pKa = 10.16FLFSLAIADD59 pKa = 4.27LLVSLFVMPMGAIPAFLGYY78 pKa = 9.61WPLGFTWCNIYY89 pKa = 8.07VTCDD93 pKa = 3.17VLACSSSILHH103 pKa = 5.91MCFISLGRR111 pKa = 11.84YY112 pKa = 9.06LGIRR116 pKa = 11.84NPLGSRR122 pKa = 11.84HH123 pKa = 6.34RR124 pKa = 11.84STKK127 pKa = 9.86RR128 pKa = 11.84LAGIKK133 pKa = 9.64IAIVWVMAMMVSSSITVLGLVNKK156 pKa = 9.48HH157 pKa = 6.27NIMPEE162 pKa = 3.8PNICMINNRR171 pKa = 11.84AFWVFGSLVAFYY183 pKa = 10.53IPMLMMVTTYY193 pKa = 11.64ALTIPLLRR201 pKa = 11.84KK202 pKa = 9.14KK203 pKa = 10.66ARR205 pKa = 11.84FAAEE209 pKa = 4.06HH210 pKa = 6.84PEE212 pKa = 4.03SEE214 pKa = 4.24LFRR217 pKa = 11.84RR218 pKa = 11.84LGGRR222 pKa = 11.84FTIRR226 pKa = 11.84PQHH229 pKa = 5.79SQQQLQMHH237 pKa = 6.74SSFSSNSNKK246 pKa = 9.24FLAMSDD252 pKa = 3.36SNRR255 pKa = 11.84NFNSSDD261 pKa = 3.18RR262 pKa = 11.84GGGRR266 pKa = 11.84GGDD269 pKa = 3.44NSTEE273 pKa = 3.96RR274 pKa = 11.84PLMQQRR280 pKa = 11.84TTSNRR285 pKa = 11.84SMGAGSVSFRR295 pKa = 11.84NVTNGGRR302 pKa = 11.84TAFGVGMGAGGRR314 pKa = 11.84SSFRR318 pKa = 11.84FAGGSILRR326 pKa = 11.84QSSTPTTTSTSLSSSCPSSTATSVRR351 pKa = 11.84GGRR354 pKa = 11.84SQQTSSFWRR363 pKa = 11.84KK364 pKa = 9.57HH365 pKa = 4.85GGNPNLMDD373 pKa = 3.28SHH375 pKa = 6.74PNRR378 pKa = 11.84RR379 pKa = 11.84ASVRR383 pKa = 11.84VSMSQPQLGYY393 pKa = 7.52PTNGNGGNGGGIGGNEE409 pKa = 4.26SPTAVTSSSCTTPGSISNLVNVATIQGQSMDD440 pKa = 3.38NHH442 pKa = 5.53QLEE445 pKa = 5.09HH446 pKa = 6.09GTTMTATITSAATTAASASARR467 pKa = 11.84ATTTTTGEE475 pKa = 4.1RR476 pKa = 11.84RR477 pKa = 11.84HH478 pKa = 6.08KK479 pKa = 10.53LRR481 pKa = 11.84PFKK484 pKa = 10.6FALNRR489 pKa = 11.84VATPTLNLRR498 pKa = 11.84FLNNRR503 pKa = 11.84NKK505 pKa = 10.44RR506 pKa = 11.84NSLSANAVATEE517 pKa = 3.99QKK519 pKa = 8.69ATKK522 pKa = 10.48VLGLVFFTFVLCWSPFFILNIIFAACPQCQVSEE555 pKa = 4.41HH556 pKa = 5.38VVNTCLWLGYY566 pKa = 10.09VSSTINPIIYY576 pKa = 9.08TIFNRR581 pKa = 11.84TFRR584 pKa = 11.84AAFIRR589 pKa = 11.84LLKK592 pKa = 10.36CNCEE596 pKa = 3.68RR597 pKa = 11.84SGRR600 pKa = 11.84PLRR603 pKa = 11.84FRR605 pKa = 11.84SVTEE609 pKa = 3.85GRR611 pKa = 11.84SALSLCAPSALPLAISFQGAPLMTPSTTNATPLSEE646 pKa = 4.21FRR648 pKa = 11.84GSYY651 pKa = 10.02TITDD655 pKa = 3.95DD656 pKa = 3.51EE657 pKa = 4.72CC658 pKa = 6.48
Molecular weight: 71.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
8086281 |
23 |
20907 |
545.6 |
60.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.226 ± 0.025 | 1.833 ± 0.031 |
5.293 ± 0.017 | 6.423 ± 0.04 |
3.488 ± 0.017 | 5.964 ± 0.026 |
2.651 ± 0.012 | 5.192 ± 0.016 |
5.688 ± 0.028 | 9.135 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.362 ± 0.013 | 5.016 ± 0.018 |
5.25 ± 0.031 | 5.357 ± 0.031 |
5.311 ± 0.02 | 8.181 ± 0.029 |
5.908 ± 0.025 | 5.685 ± 0.017 |
0.999 ± 0.008 | 3.032 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
