
Hawaiian green turtle herpesvirus
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Scutavirus; Chelonid alphaherpesvirus 5; Green turtle herpesvirus
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
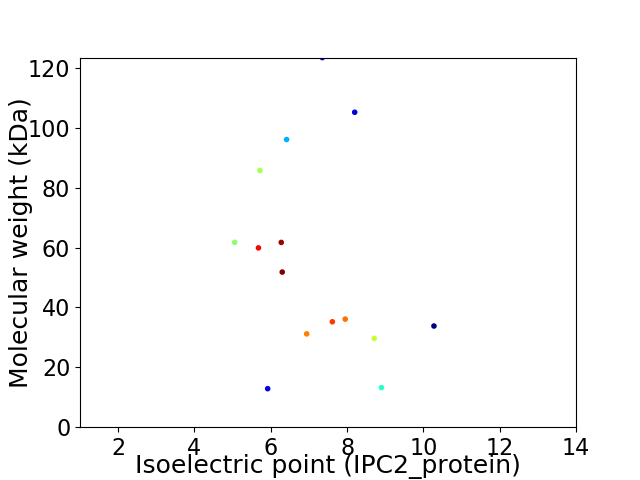
Virtual 2D-PAGE plot for 15 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|Q5ZR62|Q5ZR62_9ALPH Basic phosphorylated capsid protein OS=Hawaiian green turtle herpesvirus OX=70564 GN=UL35 PE=4 SV=1
TT1 pKa = 7.07AAGPVSEE8 pKa = 4.6TDD10 pKa = 3.29PVLRR14 pKa = 11.84SPLDD18 pKa = 3.52RR19 pKa = 11.84SEE21 pKa = 4.82LNARR25 pKa = 11.84DD26 pKa = 3.94ALLLLAAFCPWHH38 pKa = 5.71VALFARR44 pKa = 11.84LPHH47 pKa = 6.01VEE49 pKa = 3.84QLEE52 pKa = 4.66FVSKK56 pKa = 9.49TLEE59 pKa = 4.23PLLSEE64 pKa = 4.2AAVGRR69 pKa = 11.84LAVSRR74 pKa = 11.84LSGKK78 pKa = 9.74QGPRR82 pKa = 11.84LLPVLSADD90 pKa = 3.72EE91 pKa = 4.39EE92 pKa = 4.23KK93 pKa = 10.82SPRR96 pKa = 11.84VFGLRR101 pKa = 11.84LRR103 pKa = 11.84DD104 pKa = 3.19WRR106 pKa = 11.84SFQVTPSLTFPEE118 pKa = 4.07IWLQTGAAEE127 pKa = 4.2ATPSAEE133 pKa = 4.3GLRR136 pKa = 11.84EE137 pKa = 4.05LQRR140 pKa = 11.84LTDD143 pKa = 4.15DD144 pKa = 3.91GKK146 pKa = 11.16AFATLLYY153 pKa = 10.35ALAYY157 pKa = 9.77HH158 pKa = 6.59CVPPAILEE166 pKa = 4.27MLWNALRR173 pKa = 11.84PRR175 pKa = 11.84DD176 pKa = 4.21LPDD179 pKa = 3.55EE180 pKa = 4.68ADD182 pKa = 2.95ASIFYY187 pKa = 10.14RR188 pKa = 11.84QRR190 pKa = 11.84ALPGEE195 pKa = 4.24SFVVSEE201 pKa = 4.52STVALQTEE209 pKa = 4.51RR210 pKa = 11.84GEE212 pKa = 4.12IPPDD216 pKa = 3.1EE217 pKa = 4.66RR218 pKa = 11.84LWIPQARR225 pKa = 11.84ARR227 pKa = 11.84TLTVGEE233 pKa = 4.5TRR235 pKa = 11.84SKK237 pKa = 11.12APPCAFDD244 pKa = 3.38FAIFAVLTGVPLVIAATLAPGAGFSRR270 pKa = 11.84EE271 pKa = 3.88SGLFLGRR278 pKa = 11.84VLLDD282 pKa = 3.42ARR284 pKa = 11.84SGDD287 pKa = 3.5AVFDD291 pKa = 3.96EE292 pKa = 4.5VRR294 pKa = 11.84QAVSSDD300 pKa = 3.45VNFLAAIEE308 pKa = 4.64KK309 pKa = 9.89EE310 pKa = 4.18YY311 pKa = 11.05QPIEE315 pKa = 4.08NVCLKK320 pKa = 10.02EE321 pKa = 4.04QIVQISMEE329 pKa = 4.56LSRR332 pKa = 11.84QRR334 pKa = 11.84LKK336 pKa = 9.92TAAPLLVLVDD346 pKa = 3.85EE347 pKa = 4.64DD348 pKa = 4.87HH349 pKa = 7.2EE350 pKa = 4.83VTHH353 pKa = 7.07LLAEE357 pKa = 4.26NPKK360 pKa = 8.75TAEE363 pKa = 4.12KK364 pKa = 10.68EE365 pKa = 4.25LIFHH369 pKa = 6.31FRR371 pKa = 11.84VPEE374 pKa = 4.05MEE376 pKa = 4.31DD377 pKa = 3.15VRR379 pKa = 11.84IPSGAQEE386 pKa = 4.35TFLPSAPPPGDD397 pKa = 3.43PLFEE401 pKa = 4.29TALGFPPNAISYY413 pKa = 9.2EE414 pKa = 3.95RR415 pKa = 11.84DD416 pKa = 3.73GIEE419 pKa = 3.58GTLNHH424 pKa = 6.39RR425 pKa = 11.84PVYY428 pKa = 9.65EE429 pKa = 4.15KK430 pKa = 10.64RR431 pKa = 11.84PPTPEE436 pKa = 3.33IFTPPEE442 pKa = 4.03SPTTNSRR449 pKa = 11.84PRR451 pKa = 11.84WYY453 pKa = 9.73DD454 pKa = 3.21DD455 pKa = 4.6KK456 pKa = 11.49EE457 pKa = 4.47IEE459 pKa = 4.3DD460 pKa = 3.88WSFPSPPDD468 pKa = 3.26SPGFSEE474 pKa = 5.57PEE476 pKa = 3.85APPAKK481 pKa = 9.79PPVSTRR487 pKa = 11.84PIPPDD492 pKa = 3.28LVLSPRR498 pKa = 11.84DD499 pKa = 3.72AYY501 pKa = 10.04PVDD504 pKa = 3.75EE505 pKa = 5.56GGLGPIPEE513 pKa = 4.56EE514 pKa = 4.33GPLRR518 pKa = 11.84SLVRR522 pKa = 11.84RR523 pKa = 11.84SLLSLLEE530 pKa = 4.27KK531 pKa = 10.3IRR533 pKa = 11.84ALRR536 pKa = 11.84RR537 pKa = 11.84FAQKK541 pKa = 9.46TGEE544 pKa = 4.17SLLTDD549 pKa = 3.62IQRR552 pKa = 11.84IKK554 pKa = 11.06LFLRR558 pKa = 4.14
TT1 pKa = 7.07AAGPVSEE8 pKa = 4.6TDD10 pKa = 3.29PVLRR14 pKa = 11.84SPLDD18 pKa = 3.52RR19 pKa = 11.84SEE21 pKa = 4.82LNARR25 pKa = 11.84DD26 pKa = 3.94ALLLLAAFCPWHH38 pKa = 5.71VALFARR44 pKa = 11.84LPHH47 pKa = 6.01VEE49 pKa = 3.84QLEE52 pKa = 4.66FVSKK56 pKa = 9.49TLEE59 pKa = 4.23PLLSEE64 pKa = 4.2AAVGRR69 pKa = 11.84LAVSRR74 pKa = 11.84LSGKK78 pKa = 9.74QGPRR82 pKa = 11.84LLPVLSADD90 pKa = 3.72EE91 pKa = 4.39EE92 pKa = 4.23KK93 pKa = 10.82SPRR96 pKa = 11.84VFGLRR101 pKa = 11.84LRR103 pKa = 11.84DD104 pKa = 3.19WRR106 pKa = 11.84SFQVTPSLTFPEE118 pKa = 4.07IWLQTGAAEE127 pKa = 4.2ATPSAEE133 pKa = 4.3GLRR136 pKa = 11.84EE137 pKa = 4.05LQRR140 pKa = 11.84LTDD143 pKa = 4.15DD144 pKa = 3.91GKK146 pKa = 11.16AFATLLYY153 pKa = 10.35ALAYY157 pKa = 9.77HH158 pKa = 6.59CVPPAILEE166 pKa = 4.27MLWNALRR173 pKa = 11.84PRR175 pKa = 11.84DD176 pKa = 4.21LPDD179 pKa = 3.55EE180 pKa = 4.68ADD182 pKa = 2.95ASIFYY187 pKa = 10.14RR188 pKa = 11.84QRR190 pKa = 11.84ALPGEE195 pKa = 4.24SFVVSEE201 pKa = 4.52STVALQTEE209 pKa = 4.51RR210 pKa = 11.84GEE212 pKa = 4.12IPPDD216 pKa = 3.1EE217 pKa = 4.66RR218 pKa = 11.84LWIPQARR225 pKa = 11.84ARR227 pKa = 11.84TLTVGEE233 pKa = 4.5TRR235 pKa = 11.84SKK237 pKa = 11.12APPCAFDD244 pKa = 3.38FAIFAVLTGVPLVIAATLAPGAGFSRR270 pKa = 11.84EE271 pKa = 3.88SGLFLGRR278 pKa = 11.84VLLDD282 pKa = 3.42ARR284 pKa = 11.84SGDD287 pKa = 3.5AVFDD291 pKa = 3.96EE292 pKa = 4.5VRR294 pKa = 11.84QAVSSDD300 pKa = 3.45VNFLAAIEE308 pKa = 4.64KK309 pKa = 9.89EE310 pKa = 4.18YY311 pKa = 11.05QPIEE315 pKa = 4.08NVCLKK320 pKa = 10.02EE321 pKa = 4.04QIVQISMEE329 pKa = 4.56LSRR332 pKa = 11.84QRR334 pKa = 11.84LKK336 pKa = 9.92TAAPLLVLVDD346 pKa = 3.85EE347 pKa = 4.64DD348 pKa = 4.87HH349 pKa = 7.2EE350 pKa = 4.83VTHH353 pKa = 7.07LLAEE357 pKa = 4.26NPKK360 pKa = 8.75TAEE363 pKa = 4.12KK364 pKa = 10.68EE365 pKa = 4.25LIFHH369 pKa = 6.31FRR371 pKa = 11.84VPEE374 pKa = 4.05MEE376 pKa = 4.31DD377 pKa = 3.15VRR379 pKa = 11.84IPSGAQEE386 pKa = 4.35TFLPSAPPPGDD397 pKa = 3.43PLFEE401 pKa = 4.29TALGFPPNAISYY413 pKa = 9.2EE414 pKa = 3.95RR415 pKa = 11.84DD416 pKa = 3.73GIEE419 pKa = 3.58GTLNHH424 pKa = 6.39RR425 pKa = 11.84PVYY428 pKa = 9.65EE429 pKa = 4.15KK430 pKa = 10.64RR431 pKa = 11.84PPTPEE436 pKa = 3.33IFTPPEE442 pKa = 4.03SPTTNSRR449 pKa = 11.84PRR451 pKa = 11.84WYY453 pKa = 9.73DD454 pKa = 3.21DD455 pKa = 4.6KK456 pKa = 11.49EE457 pKa = 4.47IEE459 pKa = 4.3DD460 pKa = 3.88WSFPSPPDD468 pKa = 3.26SPGFSEE474 pKa = 5.57PEE476 pKa = 3.85APPAKK481 pKa = 9.79PPVSTRR487 pKa = 11.84PIPPDD492 pKa = 3.28LVLSPRR498 pKa = 11.84DD499 pKa = 3.72AYY501 pKa = 10.04PVDD504 pKa = 3.75EE505 pKa = 5.56GGLGPIPEE513 pKa = 4.56EE514 pKa = 4.33GPLRR518 pKa = 11.84SLVRR522 pKa = 11.84RR523 pKa = 11.84SLLSLLEE530 pKa = 4.27KK531 pKa = 10.3IRR533 pKa = 11.84ALRR536 pKa = 11.84RR537 pKa = 11.84FAQKK541 pKa = 9.46TGEE544 pKa = 4.17SLLTDD549 pKa = 3.62IQRR552 pKa = 11.84IKK554 pKa = 11.06LFLRR558 pKa = 4.14
Molecular weight: 61.74 kDa
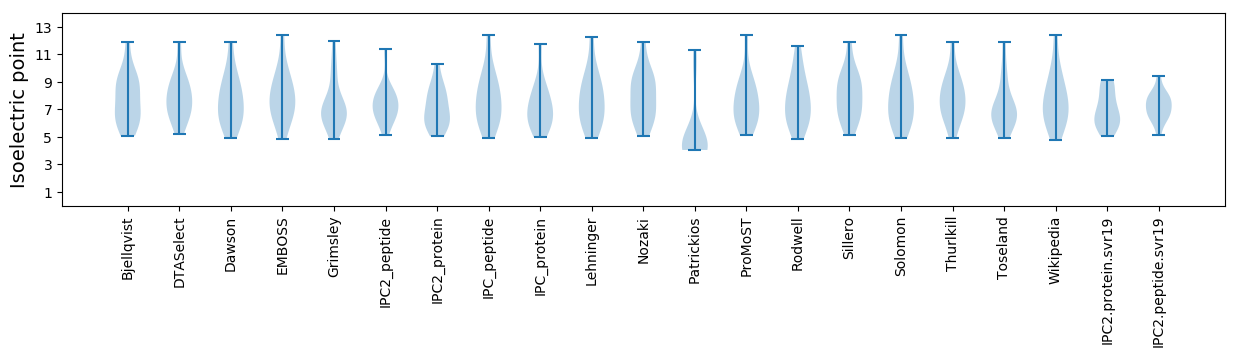
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q5ZR74|Q5ZR74_9ALPH Thymidine kinase (Fragment) OS=Hawaiian green turtle herpesvirus OX=70564 GN=UL23 PE=3 SV=2
MM1 pKa = 6.76NTLRR5 pKa = 11.84TRR7 pKa = 11.84GARR10 pKa = 11.84TSAFDD15 pKa = 3.7YY16 pKa = 9.7TSSRR20 pKa = 11.84AVKK23 pKa = 9.99KK24 pKa = 10.29RR25 pKa = 11.84RR26 pKa = 11.84LLAGVRR32 pKa = 11.84CHH34 pKa = 5.96EE35 pKa = 4.12RR36 pKa = 11.84FYY38 pKa = 11.8NLLTRR43 pKa = 11.84GSQQINAGRR52 pKa = 11.84PGAAAARR59 pKa = 11.84PLVRR63 pKa = 11.84ALGAAVDD70 pKa = 4.1CRR72 pKa = 11.84RR73 pKa = 11.84EE74 pKa = 3.91IIKK77 pKa = 9.52TGVICEE83 pKa = 3.98IDD85 pKa = 3.58LGPRR89 pKa = 11.84RR90 pKa = 11.84PDD92 pKa = 2.99CVIYY96 pKa = 10.44VRR98 pKa = 11.84RR99 pKa = 11.84ASPVPHH105 pKa = 6.98LLFLVEE111 pKa = 5.36LKK113 pKa = 10.77SCLHH117 pKa = 5.7TRR119 pKa = 11.84DD120 pKa = 3.01MATRR124 pKa = 11.84TKK126 pKa = 10.77ALQRR130 pKa = 11.84QEE132 pKa = 4.61GLGQLRR138 pKa = 11.84DD139 pKa = 3.46ARR141 pKa = 11.84ACLGRR146 pKa = 11.84LTPPGPEE153 pKa = 4.13PLLICPVLIFVAQKK167 pKa = 10.32GLRR170 pKa = 11.84ILAVQRR176 pKa = 11.84LPTVAASVHH185 pKa = 5.87FDD187 pKa = 2.94RR188 pKa = 11.84LLRR191 pKa = 11.84VLLGRR196 pKa = 11.84SCYY199 pKa = 9.85HH200 pKa = 5.91VRR202 pKa = 11.84TAPGSPLDD210 pKa = 3.9ARR212 pKa = 11.84SQRR215 pKa = 11.84SGARR219 pKa = 11.84GVAQPAKK226 pKa = 9.58PAAPSKK232 pKa = 10.58LPSIPRR238 pKa = 11.84SRR240 pKa = 11.84SNPRR244 pKa = 11.84SAGPRR249 pKa = 11.84RR250 pKa = 11.84SLAGQRR256 pKa = 11.84RR257 pKa = 11.84AARR260 pKa = 11.84PKK262 pKa = 9.03VRR264 pKa = 11.84GAGSHH269 pKa = 4.78GTAGGKK275 pKa = 9.58HH276 pKa = 5.77FDD278 pKa = 3.9HH279 pKa = 7.8RR280 pKa = 11.84SGSNNAAGRR289 pKa = 11.84AGRR292 pKa = 11.84GRR294 pKa = 11.84FKK296 pKa = 10.86NRR298 pKa = 11.84RR299 pKa = 11.84GPGSRR304 pKa = 11.84AGGRR308 pKa = 11.84ALPANN313 pKa = 4.1
MM1 pKa = 6.76NTLRR5 pKa = 11.84TRR7 pKa = 11.84GARR10 pKa = 11.84TSAFDD15 pKa = 3.7YY16 pKa = 9.7TSSRR20 pKa = 11.84AVKK23 pKa = 9.99KK24 pKa = 10.29RR25 pKa = 11.84RR26 pKa = 11.84LLAGVRR32 pKa = 11.84CHH34 pKa = 5.96EE35 pKa = 4.12RR36 pKa = 11.84FYY38 pKa = 11.8NLLTRR43 pKa = 11.84GSQQINAGRR52 pKa = 11.84PGAAAARR59 pKa = 11.84PLVRR63 pKa = 11.84ALGAAVDD70 pKa = 4.1CRR72 pKa = 11.84RR73 pKa = 11.84EE74 pKa = 3.91IIKK77 pKa = 9.52TGVICEE83 pKa = 3.98IDD85 pKa = 3.58LGPRR89 pKa = 11.84RR90 pKa = 11.84PDD92 pKa = 2.99CVIYY96 pKa = 10.44VRR98 pKa = 11.84RR99 pKa = 11.84ASPVPHH105 pKa = 6.98LLFLVEE111 pKa = 5.36LKK113 pKa = 10.77SCLHH117 pKa = 5.7TRR119 pKa = 11.84DD120 pKa = 3.01MATRR124 pKa = 11.84TKK126 pKa = 10.77ALQRR130 pKa = 11.84QEE132 pKa = 4.61GLGQLRR138 pKa = 11.84DD139 pKa = 3.46ARR141 pKa = 11.84ACLGRR146 pKa = 11.84LTPPGPEE153 pKa = 4.13PLLICPVLIFVAQKK167 pKa = 10.32GLRR170 pKa = 11.84ILAVQRR176 pKa = 11.84LPTVAASVHH185 pKa = 5.87FDD187 pKa = 2.94RR188 pKa = 11.84LLRR191 pKa = 11.84VLLGRR196 pKa = 11.84SCYY199 pKa = 9.85HH200 pKa = 5.91VRR202 pKa = 11.84TAPGSPLDD210 pKa = 3.9ARR212 pKa = 11.84SQRR215 pKa = 11.84SGARR219 pKa = 11.84GVAQPAKK226 pKa = 9.58PAAPSKK232 pKa = 10.58LPSIPRR238 pKa = 11.84SRR240 pKa = 11.84SNPRR244 pKa = 11.84SAGPRR249 pKa = 11.84RR250 pKa = 11.84SLAGQRR256 pKa = 11.84RR257 pKa = 11.84AARR260 pKa = 11.84PKK262 pKa = 9.03VRR264 pKa = 11.84GAGSHH269 pKa = 4.78GTAGGKK275 pKa = 9.58HH276 pKa = 5.77FDD278 pKa = 3.9HH279 pKa = 7.8RR280 pKa = 11.84SGSNNAAGRR289 pKa = 11.84AGRR292 pKa = 11.84GRR294 pKa = 11.84FKK296 pKa = 10.86NRR298 pKa = 11.84RR299 pKa = 11.84GPGSRR304 pKa = 11.84AGGRR308 pKa = 11.84ALPANN313 pKa = 4.1
Molecular weight: 33.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7492 |
111 |
1091 |
499.5 |
55.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.797 ± 0.568 | 1.962 ± 0.265 |
4.979 ± 0.335 | 6.887 ± 0.439 |
4.805 ± 0.241 | 6.1 ± 0.377 |
1.895 ± 0.162 | 2.803 ± 0.249 |
3.991 ± 0.253 | 10.918 ± 0.601 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.522 ± 0.165 | 3.217 ± 0.359 |
6.313 ± 0.814 | 3.31 ± 0.193 |
8.649 ± 0.539 | 6.38 ± 0.375 |
5.539 ± 0.294 | 6.874 ± 0.37 |
0.921 ± 0.127 | 3.137 ± 0.256 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
